|
|
Post by Admin on Aug 27, 2013 15:44:20 GMT
 The Udmurts in the Udmurt Republic with a high frequency (12.7%) of haplogroup D (Bermisheva et al. 2002). The second most common haplogroup in all northern Asian populations is haplogroup D, which is also very common in eastern, central Asia and America. Haplogroup D encompasses almost 20% of the total mtDNA variation in most of northern Asia and retains a very high overall frequency in all regional northern Asian groups (11–34%), central Asian (14–20%) and eastern Asian (10–43%) populations (Table S2). Its frequency declines towards the west and south, to 2% or less in India and western Asia, but in the Caucasus, Volga-Ural Region and southeastern Asia is still as high as 5–10%. Interestingly, haplogroup D is also found in some northeastern Europeans, like Karelians, Saami and Scandinavians, while haplogroup C is absent among them (Table S2). From the coalescence analysis it is evident that besides D4b1a2, only two other clusters bear the strongest signal for the post LGM expansion in northern Asia. Subclusters D4m2 and D2 demonstrate a coalescence age of 12–20 kya and 11–15 kya, respectively, which are comparable with the age of D4b1a2. It is also remarkable that within D4m2, an Altaian branch precedes subcluster D4m2a, which is characteristic for a broad range of Arctic, Subarctic and southern Siberian populations (Figure S2). Another D4 subcluster, D2, has its most likely homeland in the Baikal region of southern Siberia, from where it expanded in the Holocene northward to northeastern Asia and further to northern America. The remaining northern Asian-specific clusters of haplogroup D are significantly younger with the age estimates not exceeding 5–8 kya (Figure 3). Among these, subclusters D4e4a and D4l2 are characterized by prevalence in the Subarctic and Arctic regions, being found mostly in Evenks and Yukaghirs, whereas several newly described subclusters within haplogroup D4j (D4j4, D4j5, D4j7, D4j8, D4j9, D4j10) demonstrate more southern geographic distribution, being detected in a variety of southern Siberian populations (Figure S2). It should be noted that the rare subcluster D4e4b has been detected in eastern Europe (in Tatars and Russians), thus pointing to a limited maternal gene flow between eastern Asia/southern Siberia and eastern Europe. One more mtDNA subcluster which may be indicative of eastern Asian influx into gene pool of eastern Europeans has been revealed in haplogroup D5a. It has been shown earlier that D5a mtDNAs, with the specific control region motif 16126-16136-16360, are present at a very low frequency in several populations of northeastern Europe (Saami, Karelians, Finns, Estonians, Komi, Russians of Arkhangelsk and Novgorod regions) as well as in central Asian Tajiks and Siberian Altaians and Mansi [10], [27], [30], [31]. Analysis of complete mtDNA phylogeny indicates that these mtDNAs belong to subhaplogroup named D5a3 defined by the only transition at np 16360 (Figure S2). It is obvious that mitochondrial genomes of Russian, Mansi and FamilyTreeDNA project individual belong to D5a3a branch harboring the entire HVS1 motif, whereas Korean mtDNA represents another D5a3 branch. In fact, this most ancestral sequence indicates that D5a3 lineages could have probably arise in eastern Asia about 16 kya, and that the other lineages, belonging to the D5a3a subgroup participated in a more recent European expansion around 2.6–3.5 kya (Figure 3). It should be noted that dispersal of Saami-specific Z1a mtDNAs shared a common ancestry with lineages from the Volga-Ural region as recently as ~3 kya probably chronicles the same. www.plosone.org/article/info:doi/10.1371/journal.pone.0015214  Haplogroup D may have come from an interbreeding event with the Neanderthals and it has been speculated that the Neanderthals were the source of haplogroup D (Evans et al. 2006). Haplogroup D2 originated in the Lake Baikal region in southern Siberia and hominid remains in Uzbekistan and in the Altai region of southern Siberia were recently found to fall within the European Neanderthal mtDNA variation (Pääbo et al. 2007). The physical similarities between the Neanderthals and the Jomon/Ainu people may prove the Neanderthal lineage of haplogroup D. Speculation about the identity of the archaic Homo population from which the microcephalin D allele introgressed into the modern human gene pool points to the Neanderthal lineage as a potential (although by no means only) candidate. Furthermore, the worldwide frequency distribution of the D allele, exceptionally Anatomically modern humans and Neanderthals shared a long period of coexistence, from as early as 130,000 years ago in the Middle East (39) to as late as 35,000 years ago in Europe (40), consistent with the estimated introgression time of the microcephalin D allele at or sometime before ≈37,000 years ago. Furthermore, the worldwide frequency distribution of the D allele, exceptionally high outside of Africa but low in sub-Saharan Africa (29), suggests, but does not necessitate, admixture with an archaic Eurasian population. Finally, our estimate of the separation time between D and non-D alleles (i.e., ≈1,100,000 years with a lower-bound confidence interval of ≈530,000 years) is largely consistent with the divergence time between modern humans and Neanderthals based on mitochondrial DNA (mtDNA) sequence difference (320,000–740,000 years; refs. 41 and 42) and with the earliest appearance of Neanderthals in the fossil record ≈500,000 years ago (43). It would be of great interest to sequence the microcephalin locus in Neanderthals or other archaic Homo lineages, should it become technically feasible to retrieve and analyze nuclear DNA from ancient hominid remains. Our results not only provide genetic evidence in support of the possibility of admixture between modern humans and an archaic Homo lineage but also support the notion that the biological evolution of modern humans might have benefited from the contribution of adaptive alleles from our archaic relatives. www.pnas.org/content/103/48/18178.full The partial skeleton of an 8–10-year-old child discovered in the late 1930s in Teshik-Tash Cave, Uzbekistan, is generally accepted to represent the easternmost extent of the Neanderthal range3. However, its Neanderthal affinities have been disputed4,5. Further to the east in the Altai region of Siberia, human remains have been found in association with Mousterian lithic technology, which is usually associated with Neanderthals in Europe but is also found in association with modern humans in the Near East and northern Africa6. To determine whether the Teshik Tash and Okladnikov individuals are genetically affiliated with European Neanderthals, we attempted to retrieve mtDNA from the left femur of Teshik Tashand the three fragmentary long bones from Okladnikov. So far,mtDNA sequences have been determined from 13 Neanderthals in Europe 11–20. Comparison of these DNA sequences with those of mtDNAs from contemporary humans shows that the Neanderthal mtDNA gene pool was distinct from that of modern humans. www.academia.edu/486419/Neanderthals_In_Central_Asia_and_Siberia About 51.8% of paternal lineages of the Japanese people belong to haplogroup O, and mostly the subgroups O3 and O2b, but what really sets them apart from other ethnic groups is another Y haplogroup D2, which makes up 35% of the Japanese male lineages. D2 is unique in Japan and the Ainu are known to have exclusively D2 (85.5%) and it's assumed that the D2 lineage is derived from the Ainu/Jomon people while haologrpup O is the genetic marker of their Yayoi ancestors from the Korean Peninsula. Geographic distribution of lineages explained the great contribution of Yayoi in our results. Hammer et al. investigated geographic distribution of Y lineages in Japanese populations. Haplogroup frequencies of the Y lineages showed U-shape cline with significant correlation with geographic distance of the populations from Kyushu. In briefs, the frequency of D2 lineage increased with increase of the distance meanwhile frequencies of O lineages decreased6. The O lineages were recognized as a Yayoi founding lineage and D2 lineage was believed to be Jomon specific6, 23. Therefore, the pattern of geographic distribution of lineages supported published archeological and anthropological results about population expansion during Jomon and Yayoi period in Japan. The archeological studies suggested general demographic density was significantly greater in eastern Japan compared to western Japan around the 3,300 years BP and a rapid increase first happened in West Japan around 2,000 years BP24. The studies of physical anthropology on human skeleton showed the new continental immigrants in West Japan, Yayoi people, have better capability to achieve enough foods to feed more people than Jomon24, 25. The pattern of population expansion may explain the great genetic contribution (about 60–72%) of Yayoi in extent Japanese. Size of continental immigration was not necessary to be very large but descendants of the immigrants increased rapidly and subsequently dispersed from West Japan to other regions. Population admixture between the continental descendants (Yayoi) and Jomon descendants shaped genetic pattern of extent Japanese. Straits between Japanese islands and Asian mainland may not act as effective barriers to the genetic admixture. www.nature.com/srep/2012/120405/srep00355/full/srep00355.html |
|
|
|
Post by Admin on Aug 27, 2013 22:44:23 GMT
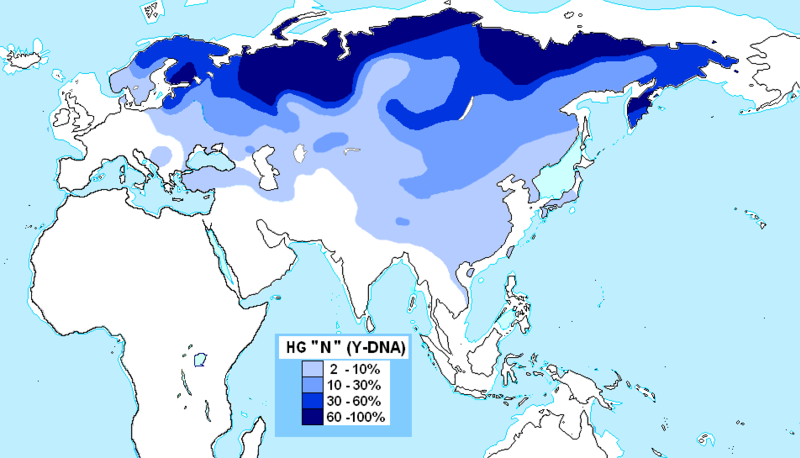 Haplogroup N (Y-DNA) is found from Finland to Siberia at high frequencies among modern Finns (58%), Lithuanians (42%), Latvians (38%), Estonians (34%), Swedes (14.4%) and northern Russians and it's also found at the highest frequency in Siberia (38.27%) and at minor frequencies in Japan (7.7%) (Hammer et al. 2005). North European populations exhibit differing amounts of IBS similarity to East Asians and Finns, especially Eastern Finns, are the most similar as confirmed by cultural and linguistic ties (i.e. whaling) and within Sweden, Norrland show the most of East Asian similarity and Götaland the least, indicating that Norrland was originally colonised by North Asians from Siberia and they were culturally and genetically integrated into Norse society in a gradual process. Haplogroup N is believed to have originated in Southeast Asia approximately 15,000 to 20,000 years ago and the N1c1 subclade in Europe likely arose in Southern Siberia 10,000 years ago, and spread to North-East Europe 6,000 years ago. Furthermore, the high frequencies of haplogroup N3 (35%) among the northern Russian population point to ancient admixture with Altaic people as Scandinavian colonists gradually spread throughout Russia.  Khanty girls  Obsko-Ugric People at Moscow exhibition N1b is the predominant haplogroup in the Khanty population; however, Y-STR variance values are much higher for both the Izhemski and Priluzski Komi groups (0.098 versus 0.181 and 0.611, respectively) (Table 4). Similarly, time estimates for the Khants reveal a rather recent entrance of the haplogroup into the population (4.0±2.6), whereas much later dates are obtained for the Izhemski (6.7±4.2) and Priluzski (12.9±4.1) Komi populations (Table 4). Variance calculations for the Pinega and Mezen populations yield Vp values of 0.163 and 0.083, respectively. Conversely, the other Slavs group attains a variance value of 0.653 and age estimate of 18.1±6.4; however, a bipartite structure is observed with separate clusters that attain ages of 6.0 ± 3.7 and 6.0±3.2. A Network Analysis, including Khanty, and the Uralic and Slavic Russian groups at a resolution of 15 Y-STR loci, displays a clear partition between the Slavic groups and the Khanty collection (Supplementary Figure 3b). Interestingly, the Izhemski Komi partitions to the portion of the projection encompassing the Slavic groups while the Komi from Priluzski shares haplotypes with both clusters. Haplogroup N is found throughout North-Central Eurasia at varying frequencies with sub-haplogroup N1c being the most widespread.15 Proposed migratory routes based on Y-STR variance estimations have suggested that N1c carriers spread from northern China through Siberia to northeastern Europe.15 Sub-haplogroup N1c1 (defined by mutation M178), long believed to be restricted to Europe and to mark a recent Uralic migration into northern Europe,14 is now known to be widespread in northern China and Mongolia.16 However, Y-STR variance values from this study do not support a migratory route from the Urals to the northeastern Slavic domain, as Russian Slavic populations exhibit higher Y-STR diversity (as high as 0.226 in the Arkhangelski population) than those found in the Uralic groups (0.079 in the Izhemski Komi and 0.121 in the Priluzski Komi) (Table 4). 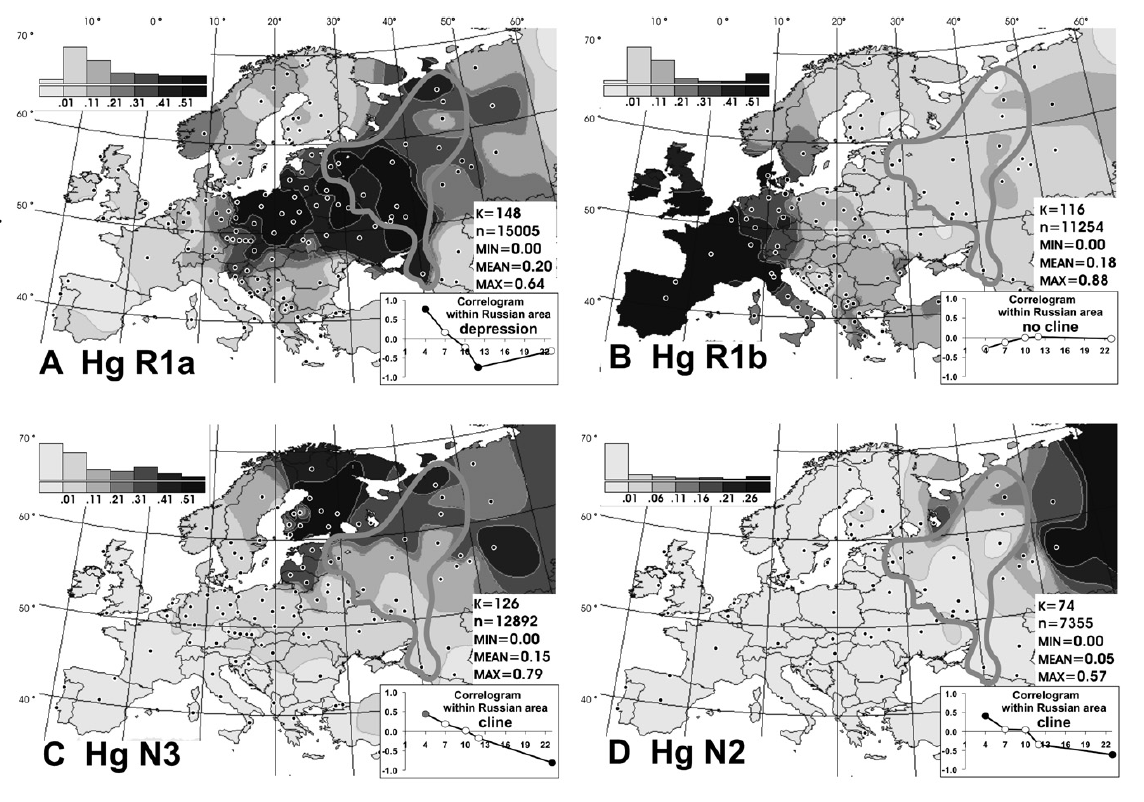 The second frequent among Russians is haplogroup N3 The second frequent among Russians is haplogroup N3 (Figure 3C), which is a typical haplogroup for Altaic and Finno-Ugric populations of Siberia and northeastern Europe.21–23 Figure 3C illustrates the fact that within the Russian area, the frequency of N3 decreases significantly from north (>35%) to south (<10%). Thus, N3 follows a trend, opposite to that observed for R1a. Similarly, haplogroup N2 reveals a northeast-to-southwest declining frequency pattern (Figure 3D). This sister group to N3 is widespread in west Siberia23,24 and is present also in Volga-Uralic region populations with frequency at approximately 20%.6,21 Northern Russians possess this haplogroup at variable frequencies (14%, 7%, and 3% in the three northern populations), whereas it is virtually absent elsewhere among Russians. Correlograms on Figures 3C and 3D strongly support clinal variation of haplogroups N2 and N3 in Russian populations. 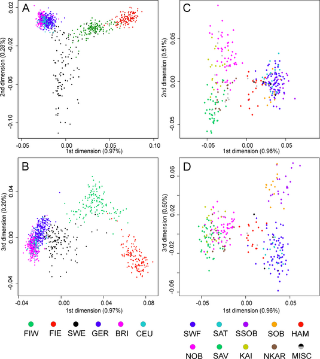 Multidimensional scaling plots of the identity by state matrices. Plots for the Europeans in the 1st and 2nd dimensions (a), and the 1st and 3rd dimensions (b), and the Finnish samples in the 1st and 2nd dimensions (c), and the 1st and 3rd dimensions (d). The label of each axis shows the proportion of the dimension’s eigenvalue to the sum of absolute eigenvalues of all dimensions: Multidimensional scaling plots of the identity by state matrices. Plots for the Europeans in the 1st and 2nd dimensions (a), and the 1st and 3rd dimensions (b), and the Finnish samples in the 1st and 2nd dimensions (c), and the 1st and 3rd dimensions (d). The label of each axis shows the proportion of the dimension’s eigenvalue to the sum of absolute eigenvalues of all dimensions: : Western Finland (FIW); Eastern Finland (FIE); Sweden (SWE); Germany (GER); Great Britain (BRI); Utah residents with ancestry from northern and western Europe (CEU); Yoruba from Ibadan, Nigeria (YRI); Han Chinese from Beijing, China (CHB); and Japanese from Tokyo, Japan (JPT). Abbreviations within Finland: Southwest Finland (SWF); Satakunta (SAT); Häme (HAM); Southern Ostrobothnia (SOB); Swedish-speaking Ostrobothnia (SSOB); Savo (SAV); Northern Karelia (NKAR); Kainuu (KAI); Northern Ostrobothnia (NOB); Miscellaneous (MISC). When data from HapMap Han Chinese+Japanese and Yoruba individuals was included in the analysis, the MDS plot of IBS formed a triangle of the three continents in the first two dimensions, with the third dimension separating the European populations clinally from each other (Fig. S3). In the histograms of IBS between the five European populations and each HapMap population (Fig. 4a), the studied populations were most similar with the CEU and least similar with YRI. Interestingly, the similarity with the Asians varied between populations, being higher for Eastern Finns, Western Finns and Swedes than for the Germans and British (p<10−14 for all comparisons except for GER and BRI whose distributions did not differ). The same pattern was also observed when comparing the allele frequencies in the study populations and in CEU and CHB+JPT: the Eastern Finns had the largest proportion of SNPs deviating towards the Asian frequencies (Table S2; p<10−5), also when markers with smallest differences were excluded (data not shown). Another factor behind the outlier status of Finland could be admixture with other populations outside the studied region. Indeed, the comparison to the Asian HapMap samples revealed interesting differences between the studied populations, with the Nordic populations and especially Eastern Finns appearing to harbour a significantly stronger Asian affinity than Central Europeans. A similar eastern influence has been observed in Y-chromosomal, mitochondrial DNA and autosomal studies of the Finns [5], [20]–[23], consistently with archaeological and linguistic data. A small degree of Saami admixture has been observed among the Finns [41] and could also contribute to the differentiation observed in this study, but it could not be detected in the absence of reference data. Thus, the possible eastern contribution observed among the Finns supports the earlier studies done with a more limited number of markers, although a full synthesis of past migration waves is beyond the scope of this study and would require additional data. www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0003519 The Y-chromosome haplogroup N-M231 (Hg N) is distributed widely in eastern and central Asia, Siberia, as well as in eastern and northern Europe. Previous studies suggested a counterclockwise prehistoric migration of Hg N from eastern Asia to eastern and northern Europe. However, the root of this Y chromosome lineage and its detailed dispersal pattern across eastern Asia are still unclear. We analyzed haplogroup profiles and phylogeographic patterns of 1,570 Hg N individuals from 20,826 males in 359 populations across Eurasia. We first genotyped 6,371 males from 169 populations in China and Cambodia, and generated data of 360 Hg N individuals, and then combined published data on 1,210 Hg N individuals from Japanese, Southeast Asian, Siberian, European and Central Asian populations. The results showed that the sub-haplogroups of Hg N have a distinct geographical distribution. The highest Y-STR diversity of the ancestral Hg N sub-haplogroups was observed in the southern part of mainland East Asia, and further phylogeographic analyses supports an origin of Hg N in southern China. Combined with previous data, we propose that the early northward dispersal of Hg N started from southern China about 21 thousand years ago (kya), expanding into northern China 12–18 kya, and reaching further north to Siberia about 12–14 kya before a population expansion and westward migration into Central Asia and eastern/northern Europe around 8.0–10.0 kya. This northward migration of Hg N likewise coincides with retreating ice sheets after the Last Glacial Maximum (22–18 kya) in mainland East Asia. Based on the dating of the Hg N haplotypes and their geographic distributions paired with the suggested counter-clock-wise migratory route across Eurasia [3], we proposed a migratory map (Figure 4) of the Hg N lineages beginning in southern China about 21 kya, and expanding into northern China 12–18 kya, reaching further north to Siberia about 12–14 kya [3], and followed by a population expansion and westward migration into Central Asia and East/North Europe around 8.0–10.0 kya [16]. www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0066102 |
|
|
|
Post by Admin on Aug 28, 2013 5:17:01 GMT
 The farmer UWS5 lineage associated with subclade N1a1a3 which is widespread in Italy, Yemen, Arabian Peninsula, Austria, Germany, Slovakia, Sweden, and Norway (Figure (Figure2c)2c) [15,20,21]. So, based on the current distribution of subcluster N1a1a3 haplotypes, it is rather difficult to establish the exact geographic origin of the farmer sample UWS5. Nevertheless, the preferential distribution of this haplotype in the Near East and central/western Europe as well as its scarce observation in eastern Europe suggests that the lineage ancestral to UWS5 dispersed probably from the Near East through the southern Europe and then into central/western Europe. However, the clarification of the most likely place of origin of farmer UWS5 lineage requires further study. The farmer lineages ECS1 and DEB1 belong to sub-branch N1a1b which is found in the Arabian Peninsula, Armenia, and Italy [3,14,20,23] (Figure (Figure2d).2d). Thus, the presence of this lineage in central Europe may represent a Near Eastern influence due to both a high frequency and a high degree of diversity of this lineage in the Arabian Peninsula. The ancestral haplotypes to European N1a1 (haplotypes with 16147G) are more common in the Arabian Peninsula, northern Africa [3,20,23-26], with a limited expansion around the Iran, Israel, Turkey, Greece [13,19,27], and are relatively rare in Europe. The coalescence time for the whole N1a haplogroup based on synonymous mutations rates as well as coding region estimates was between 19,600-23,500 years (Table (Table2).2). Overall the distinct phylogeographic distribution of N1a subclusters and their coalescence times suggests that an initial diversification of N1a occurred in the Near East, followed by westwards dispersion of ancestors of particular subhaplogroup to southern Europe and northwards via Central Asian steppe zones to central Europe. Based on the current N1a haplogroup phylogeny and phylogeographic information on the farmer mtDNA associated subclades distribution, we suppose that the farmer lineages-DEB3, FLO1, and HAL2 might be derived from local communities and that they would have adopted the farming culture indigenously. Therefore, the results of the present study are somewhat difficult to reconcile with the hypothesis that the N1a lineages were brought into central Europe by the Neolithic farmers from the Near East by a major demic diffusion event. Moreover, the evidence from phylogeographic analysis of N1a lineages emphasizes that European farmer N1a lineages might have been originated from different sources- from eastern Europe (for N1a1a1), from Near East via southern Europe (for N1a1b and perhaps for N1a1a3), and from local central European source (for N1a1a2). It is thus clear that Neolithic farmers' migration into central Europe did not occur in a uniform way; indeed these results indicate that the Neolithic transition process was more complex in central Europe and possibly the farmer N1a lineages were brought in through the 'leapfrog' colonization process [5,28]. www.ncbi.nlm.nih.gov/pmc/articles/PMC2964711/ |
|
|
|
Post by Admin on Aug 29, 2013 5:11:34 GMT
 Using PCA analysis, the haplogroups found in the Swedish population sample agreed with haplogroup distributions in other European countries (data not shown). The most common haplogroup in Sweden was I1a*, which is also present in the same frequency in Norway.27 I1a* is thought to have a decreasing gradient from Scandinavia towards both the east (Ural) and the west (Atlantic) and Western Europe has been suggested as the source of the Scandinavian I1a*.27 Semino et al28 concluded that haplogroups with the M170 mutation (defining haplogroup I) and the M173 mutation (defining haplogroup R1*) have been present in Europe since the palaeolithic period, while the other haplogroups entered, independently, from the Middle East and the Urals. R1b3 and R1a1 (24 and 12% in Sweden respectively) are today two common haplogroups in the rest of Europe. It has been suggested that the mutation determining R1*(xR1a1) originated 35 000–40 000 years ago in western Europe and that the mutation defining R1a1 arose later on in eastern Europe. Semino et al also suggested that these two haplogroups expanded into the central parts of the continent after the last glacial maximum. Studies of today exhibit that these two haplogroups show a gradient pattern in Europe. The fourth most frequent haplogroup in Sweden was N3 (10%). This haplogroup is mostly present in the northern Swedish regions, indicating a closer relationship with Saami and Finnish populations, in which N330, 31 is very common (this is discussed further below). Tambets et al31 suggested that the higher diversity found in eastern Europe (compared to Siberia) would make eastern Europe a possible origin for this haplogroup. The time for its expansion remains, however, unclear. There are differences in haplogroup frequencies among all the regions studied. Some may be due to the limited sample size. Anyhow, all analyses in this study provided information suggesting that Västerbotten differed from the rest of the Swedish regions. This deviation is mostly due the high frequencies of N3 and I1c haplotypes compared to the other regions. N3 and I1c together account for 37% of the Y chromosomes in Västerbotten, but only 4–15% in the other Swedish regions and, at the same time, the lower frequency of I1a* and R1b3, which are very common in the other regions (40% together in Västerbotten but 60–73% in other Swedish regions). Furthermore, when N3 and I1c were excluded from the exact test for population differentiation the earlier significant difference disappeared. The high frequency of N3 in Västerbotten can be explained by the short geographical distance to both the Saami and the Finnish populations. However, our study suggests a closer relationship (represented by pairwise RST values) between Saami and Västerbotten N3 haplotypes than between Västerbotten and Österbotten haplotypes. Among the Österbotten N3 Y chromosomes, 13 of 26 haplotypes were identical (14-14-30-24-11-14-14-11-13) which makes the Österbotten haplotype diversity extremely low, suggesting a high grade of endogamy or founder effects. This observation has been made in many previous studies.30, 32 The same haplotype is found three times among 17 Saami N3 Y chromosomes and in only one of the Västerbotten Y chromosomes. The absence of the common Finnish haplotype in Västerbotten may explain a more distant relationship to Österbotten, relative to the Swedish Saami population. www.nature.com/ejhg/journal/v14/n8/full/5201651a.html |
|
|
|
Post by Admin on Aug 29, 2013 21:34:29 GMT
.PNG) Haplogroup R1b is the most frequently occurring Y-chromosome haplogroup in Western Europe but it is also prevalent in some parts of Central Africa and South Asia. In northern Cameroon, R1b can be found among 95.5% of the population and R1b is likely to have originated in Central Africa and it gradually spread all across Eurasia as a group of R1b people out of Africa made its way towards Europe and Asia. Cameroon is the plausible place where haplogroup R1b came into existence and the map below shows their migration route out of Africa via Egypt. The controversial theory of a possible back migration from Asia now has been discredited (Cruciani et al. 2002) and Eupedia removed a related entry. Haplogroup R1 is the most common haplogroup throughout Eurasia from Britain (60-80%) to India (45-50%) and this recent paper (Winters 2010) confirmed haplogroup R1's African roots. A pristine form of R1 can be found at the highest frequency in northern Cameron (95.5%) and haplogroup R1 also pertains to the ancient Kushite people who moved out of the Upper Nile area into Eurasia (Genesis 10) and they had inhabited in Cameroon, the Niger Valley and Senegambian regions in ancient times prior to their migration out of Africa. Moreover, Assyrians are one of the oldest surviving groups descended,as it is believed, from the historic Sumers. Among Assyrians,R1b is the major haplogroup, reaching 40% of the studiedpopulation; haplogroup J takes second place with 11% andthers are in singular percentages (Lashgary et al. 2011). In this paper we discuss the role of the Kushites in the spread of R1*-M173. Human y-chromosomehaplogroup R1*-M173 is mainly found in Africa. Haplogroup R1*-M173 is the pristine form of haplogroup R. In Africa researchers have detected frequencies as high as 95% among Sub-Saharan Africans. The phylogenetic, craniometric, textual, historical and linguistic evidence support the demic diffusion of Niger-Congo (Nilo-Saharan) carriers of R1*-M173 from Africa to Eurasia between 4-5kya. The phylogenetic profile of R-M173 supports an ancient migration of Kushites from Africa to Eurasia as suggested by the Classical writers. In Fig. 3, we outline the spread of haplogrorp R from Nubia into Asia and West Africa. This expansion of an African Kushite population probably took place Neolithic period. The accumulated Classical literature, archaeological,craniometric, genetic and linguistic evidence suggest a genetic relationship between the Kushites of Africa and Kushites in Eurasia that cannot be explained by microevolutionary mechanisms. The phylogeographic profile of R1*-M173 supports this ancient migration of Kushites from Africa to Eurasia as suggested by the Classical writers. This expansion of Kushites into Eurasia probably took place over 4 kya. The linguistic evidence makes it clear that the Nilo-Saharan and Niger-Congo languages are related. The genetic evidence indicates that Nilo-Saharan and Niger-Congo speakers carry the y-chromosomes M3b*-M35 andR1*-M173, an indicator for the earlier presence of speakers of this languages in an original Nile Valley homeland. The distribution of y-chromosome specific haplogroups in areas formerly occupied by the Kushite people of Asia reveal continuity between the ancient inhabitants of Anatolia, Mesopotamia and Persia and Africa. The genetic pattern indicates a significant Sub-Saharan male contribution to the populations presently situated in south-western Eurasia. The tradition of a Kushite migration from Africa to Asia recorded in the classical literature is supported by the clinal biological pattern of y-chromosome lineages in Africa and Eurasia. The presence of R1*-M173 among Anatolians and Iranians supports a Neolithic demic diffusion of Kushite agropastoral populations into this region. The cranial discrete traits, y-chromosome haplogroups and linguistic affiliations shared between Sub-Saharan Africans, the ancient Mesopotamian, Anatolian and Iranian populations can only be the result of a human migration from Africa to Eurasia in ancient times as noted by the Classical writers of Greece and Rome. .PNG/300px-Haplogroup_R1b_(Y-DNA).PNG) Haplogroup R1b frequency in Europe is clinal with increasing frequencies observed in Northwest Europe, a pattern that has been ascribed to the persistance of Palaeolithic Y chromosomes in Europe after a Neolithic demic diffusion from the Near East.20, 21 Interestingly, attempts to date the Y-STR-based diversity of R1b-M269 chromosomes in populations from Europe and Turkey have yielded Holocene expansion times in both regions.7, 22, 23 These findings have led to the reappraisal that R1b-M269 in Europe is young and likely associated with a Neolithic demic expansion from the Near East through Anatolia.22, 23 Although the frequency of R1 lineages is currently the highest in Europe, the phylogeographic argument for their origin outside Europe, likely somewhere in West Asia, arises from the geographic distribution of the primary splits in the R1 phylogeny: at least three basic R-M207-derived haplogroups – R1a-M420*, R1b-M343* and R2 – occur mostly outside Europe. Figure 1b shows approximate locations of the 118 populations studied and proportional sample sizes. As the intensity of sampling is thin relative to the expanse of West Asia, the spatial-frequency surfaces for this region should be viewed as preliminary. Of the total of 193 R1b-M73 chromosomes detected, all except two Russians occurred outside Europe, either in the Caucasus, Turkey, the Circum-Uralic and North Pakistan regions (Figure 1c), in contrast to its considerably more widespread companion R1b-M269 clade (Figure 1d). With the exception of rare incidences of R1b-V88 in Corsica, Sardinia13 and Southern France (Supplementary Table S4), there is nearly mutually exclusive patterning of V88 across trans-Saharan Africa vs the prominence of P297-related varieties widespread across the Caucasus, Circum-Uralic regions, Anatolia and Europe. The detection of V88 in Iran, Palestine and especially the Dead Sea, Jordan (Supplementary Table S4) provides an insight into the back to Africa migration route. The initial arrival of farmers from Southwest Asia to the present-day Greece occurred ca 9000 years BP.38 Outside of Southeast Europe, two episodes of early farming are attested archeologically.39 The first involved a maritime colonization of Crete ca 9000 years BP and Southern Italy ca 8000 years BP and subsequently spread to coastal Mediterranean France and Spain, as exemplified by impressed/cardial pottery. The second involved a migration to Central Europe, from Hungary to France, characterized by LBK (ca 7500 years BP). Within a 3k-year period, the agricultural economy spread across Europe, terminating in Britain and Scandinavia ∼6000 years BP.39 Our results implicate complexity in the post-glacial formation and expansion of populations in Europe during the past ca 10 000 years. The narrow temporal window between potential expansions by Mesolithic foragers at the onset of the Holocene (10k years ago) and pioneer farmers from the Near East during the early Neolithic into Central Europe (7.5k years ago) is exceedingly difficult to discern with genetic tools.22 Thus, invoking the pronounced transformation of the pre-Neolithic European gene pool by intrusive pioneer farmers from the Near East must be viewed cautiously especially when such an argument is based on just a single incompletely resolved haplogroup. Although the transition to agriculture was a pivotal event in human history, the spread of specific haplogroups can occur in more than one migration event. Evidence of trade networks based on the exchange of commodities (eg salt, amber) along northwest to south and southeast directions, eg the Iron Age Hallstatt Culture,43 provided opportunities for potential gene dispersion. However, the magnitude of such putative commodity-driven gene flows remains uncertain until direct evidence from ancient DNA is provided in combination with potentially even more high-resolution and informative sub-haplogroup fractions relevant to particular trade routes or cultural horizons are detected and used to test hypotheses concerning post-Neolithic histories. www.ncbi.nlm.nih.gov/pmc/articles/PMC3039512/ |
|













.PNG/300px-Haplogroup_R1b_(Y-DNA).PNG)