|
|
Post by Admin on Sept 17, 2019 20:58:18 GMT
Nearly 2000 years ago, Rome was the largest urban center of the ancient world and the capital of an empire with over 60 million inhabitants. Although Rome has long been a subject of archaeological and historical study, little is known about the genetic history of the Roman population. To fill this gap, we performed whole genome sequencing on 127 individuals from 29 sites in and around Rome, spanning the past 12,000 years. Using allele frequency and haplotype-based genetic analyses, we show that Italy underwent two major prehistoric ancestry shifts corresponding to the Neolithic transition to farming and the Bronze Age Steppe migration, both prior to the founding of the Roman Republic. As Rome expanded from a small city-state to an empire controlling the entire Mediterranean, the city became a melting pot of inhabitants from across the empire, harboring diverse ancestries from the Near East, Europe and North Africa. Furthermore, we find that gene flow between Rome and surrounding regions closely mirrors Rome’s geopolitical interactions. Interestingly, Rome’s population remains heterogeneous despite these major ancestry shifts through time. Our study provides a first look into the dynamic genetic history of Rome from before its founding, into the modern era.  Due to their strategic geographic location between three different continents, Sicily and Southern Italy have long represented a major Mediterranean crossroad where different peoples and cultures came together over time. However, its multi-layered history of migration pathways and cultural exchanges, has made the reconstruction of its genetic history and population structure extremely controversial and widely debated. To address this debate, we surveyed the genetic variability of 326 accurately selected individuals from 8 different provinces of Sicily and Southern Italy, through a comprehensive evaluation of both Y-chromosome and mtDNA genomes. The main goal was to investigate the structuring of maternal and paternal genetic pools within Sicily and Southern Italy, and to examine their degrees of interaction with other Mediterranean populations. Our findings show high levels of within-population variability, coupled with the lack of significant genetic sub-structures both within Sicily, as well as between Sicily and Southern Italy. When Sicilian and Southern Italian populations were contextualized within the Euro-Mediterranean genetic space, we observed different historical dynamics for maternal and paternal inheritances. Y-chromosome results highlight a significant genetic differentiation between the North-Western and South-Eastern part of the Mediterranean, the Italian Peninsula occupying an intermediate position therein. In particular, Sicily and Southern Italy reveal a shared paternal genetic background with the Balkan Peninsula and the time estimates of main Y-chromosome lineages signal paternal genetic traces of Neolithic and post-Neolithic migration events. On the contrary, despite showing some correspondence with its paternal counterpart, mtDNA reveals a substantially homogeneous genetic landscape, which may reflect older population events or different demographic dynamics between males and females. Overall, both uniparental genetic structures and TMRCA estimates confirm the role of Sicily and Southern Italy as an ancient Mediterranean melting pot for genes and cultures. The first unquestioned colonization of Sicily has been linked to the Palaeolithic, and in particular to Epigravettian human groups coming from the mainland and entering Sicily through the present-day Strait of Messina [2]–[3]. Human remains, referable to the Upper Palaeolithic, recently discovered in Southern Italy (Grotta of Paglicci, Puglia [4]) and Sicily (Grotta d'Oriente in the island of Favignana, [5]), have been attributed to the mtDNA haplogroup HV and tentatively interpreted as descendants of the early-Holocene hunter-gatherers of Sicily and Southern Italy, who occupied this area before (Gravettian) and after (Epigravettian) the Last Glacial Maximum [5]. The transition to agriculture with the Neolithic revolution, occurred in the South-Eastern heel of Italy between 6000–5700 years BCE, then moving west towards Southern Calabria and Eastern Sicily, where traces of the same material cultures (imprinted ceramics stentinelliane) have been dated roughly to 5800–5400 BCE [6]. However the Neolithic pottery (imprinted ceramics prestentinelliane) uncovered in western Sicily (Uzzo and Kronio) are coeval (6000–5750 BCE) with the earliest occurrence of Neolithic materials in the more South-Eastern portion of the Italian Peninsula, thus suggesting potentially parallel and culturally independent processes of colonization between the eastern and western parts of the island [6]. 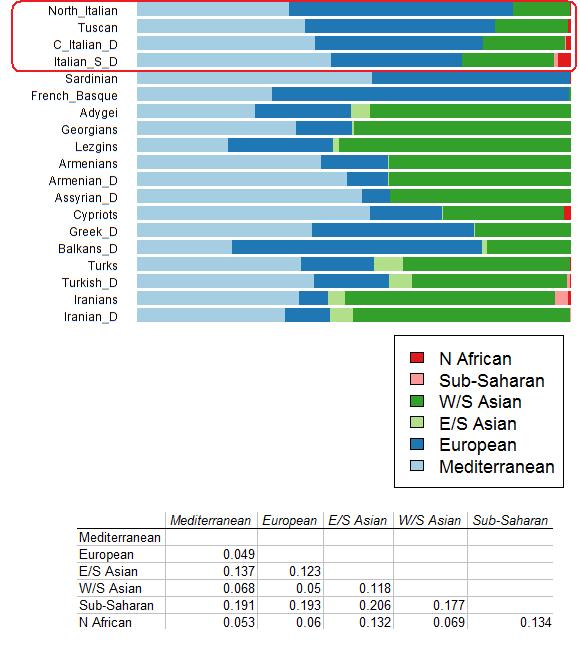 In addition to Upper-Palaeolithic and Neolithic material cultures, historical and archaeological data offer a detailed and reliable understanding of the more recent population influences on Sicily and Southern Italy. Among the well-documented historical events, at least four main migration processes could potentially have affected the current genetic variability of the area: i) the massive occupation of Greeks (giving rise to the “Magna-Graecia”) started in the 8th century BC from the Southern Balkans; ii) the Phoenician and Carthaginian colonization of the western part of Sicily occurred since the first millennium BC from the Levant through North Africa; iii) the Roman and post-Roman (Germanic) invasions from continental Italy and Central-Western Europe between the 300 BC and 500 AD; and iv) the more recent Muslim and Norman conquests of Sicily and Southern Italy in 8th–9th and 11th–12th centuries AD respectively. If on one hand the Greek colonisation of the south-eastern regions vs. the Phoenician occupation of western Sicily could have caused internal east-west cultural differentiation, on the other hand the later conquests (such as Germanic, Islamic and Norman occupations) may have contributed to reshape at different levels the genetic landscape of one of the largest Mediterranean islands, albeit their relative impacts remain still questioned. Such a deep and complex historical stratification made the reconstruction of the genetic history and population structure of the area open to debate. Previous investigations on the genetic structure of Sicily, based on both classical, autosomal and uniparental markers, have indeed shown contrasting results about the presence [7]–[8] or the absence [9] of an east-west geographically heterogeneous distribution of genetic variation within the island [8]. By contrast, a substantial homogeneity in genetic variation, emerged from recent mtDNA-based studies focused on specific regions of Southern Italy [10]–[11]. To the best of our knowledge, all previous studies that specifically addressed the reconstruction of the genetic structure and population history of Sicily and Southern Italy, have been mostly focused on only one of the two areas at a time, moreover considering the maternal (mtDNA) and the paternal (Y-chromosome) perspectives separately.  In this study we present an high-resolution analysis of the uniparental genetic variability of Sicily and Southern Italy, by using a new accurately selected set of samples and, for the first time, by jointly analysing both paternal and maternal genetic systems at the same time. More than 300 individuals from 8 different Sicilian and Southern Italian provinces have been deeply typed for 42 Y-SNPs and 17 Y-STRs, as well as for the HVS-I and HVS-II regions and 22 coding SNPs of mtDNA. These data have been used to compare and contrast Y-chromosome and mtDNA genetic patterns within Sicily and Southern Italy, and then to investigate their affinities within the overall Mediterranean genetic landscape by further comparing our data with those of reference populations selected from Central, Western and Southern Europe, as well as from North Africa and the Levant. In this way we particularly seek to address the following questions: i) Is the genetic diversity of Sicily structured along its east-west axis and how is it patterned compared to Southern Italy? ii) Are the observed genetic patterns stratified temporally or geographically in terms of more ancient or recent peopling events, and are there any differences between maternal and paternal perspectives? iii) How is the genetic variability of Sicily and Southern Italy related to the wider Euro-Mediterranean genetic space and what are the main contributes to the current genetic pool? Since Sicily and Southern Italy have long played an important key role in the history of demic and cultural transitions occurred in Southern Europe and the Mediterranean, the clarification of these points will be of great relevance for the understanding of the different population, cultural and linguistic dynamics occurred within the whole Mediterranean area. |
|
|
|
Post by Admin on Sept 17, 2019 21:31:31 GMT
Y-Chromosome perspective The 326 unrelated individuals from 8 different locations of SSI have been assigned to 33 different haplogroups whose frequencies, for both the whole dataset as well as for each of the 8 sampling points, are detailed in Table S2. Y-STR haplotypes for the 119 newly-typed individuals are provided in Table S3. Haplogroups G-P15 (12.3%), E-V13 and J-M410* (both 9.5%), together with R-M269* (7.4%) represent the most frequent lineages found in Sicily and Southern Italy (SSI). These are followed by five R1-sublineages (R-M17, R-L2, R-P312, R-U152, R-U106), whose frequencies range from 5.2% to 3.7%, and by J-M267 which embraces almost the 5% of total variability. All these paternal lineages reportedly originated in Europe or in the Near East, whereas much lower it seems to be the African paternal contribution, mainly represented by haplogroups belonging to HG-E sub-lineages (E-V12, 2.76%; E-V22, 2.15%; E-M81, 1.53%). Contrary to what previously reported in literature [8], no differential distribution of Y-chromosome lineages has been found in our dataset. Fisher exact tests performed on HG frequencies between Southern Italy and Sicily (P-value: 0.4765), as well as between Eastern and West Sicily (P-value: 0.2998), indeed do not reveal any significant differentiation. No significant percentage of variance among groups of populations (FCT) has been detected by regional AMOVAs (Table S4). In the same way, when our Sicilian populations were grouped with those of Di Gaetano et al. 2009 following their East-West subdivision scheme and by using the same HG resolution level, both AMOVA (variation among groups 0.30%, P-value 0.091) and Fst index (P-value 0.094), failed to reveal any significant difference in Y-chromosome HGs composition, thus pointing out a substantial homogeneous pattern of genetic variation within the island. Moreover, when the distribution of Y-chromosome lineages in the present-day Sicilian and Southern-Italian population has been compared with the one of the surname-based selected subset, no significant differentiation appeared (P-value: 0.9551).  Figure 1. Spatial Principal Component Analysis (sPCA) based on Y-chromosome haplogroups frequencies. The first two global components, sPC1 (a) and sPC2 (b), are depicted. Positive values are represented by black squares; negative values are represented by white squares; the size of the square is proportional to the absolute value of sPC scores. Interestingly, the second sPC (Figure 1b), despite being much less representative compared to the first one in terms of both variance and spatial autocorrelation, identifies a subdivision between the two Mediterranean coastlines, which seems to involve the Eastern and Western parts of Sicily. The first group (black squares) is indeed represented by populations from the South-Eastern Mediterranean shore (Levant and North-Africa), including also the most western Sicilian provinces (Trapani and Agrigento) and the Iberian populations. Conversely, the second cluster (white squares) is mainly a North-Eastern Mediterranean centred group, encompassing the Balkans, South-Italy and East-Sicily, together with the other central European populations. When the reliability of the sPCA-identified structures was tested by means of an AMOVA based on haplogroup frequencies, the proportion of genetic variation between groups (FCT) results however two times higher when grouping according to the sPC1 (8.31%, P-value<0.001) than sPC2 (4.31%, P-value = 0.004). The sPCA-suggested pattern of genetic relationships among the different Mediterranean populations, has been confirmed in the classical PCA plots reported in Figure S2a The two high-structured Mediterranean clusters identified with sPC1, were further tested by means of DAPC analysis. Membership probabilities, represented with a structure-like plot (Figure 2), highlight the intermediate position of the Italian samples between the two Mediterranean clusters. In this context, Sicily and Southern Italy show clearly their stronger affinity with the populations from the South-Eastern Mediterranean side (with the partial exception of Catania - CT).  Figure 2. Discriminant Analysis of Principal Components (DAPC) based on Y-chromosome sPC1-identified structure. The barplot represents DAPC-based posterior membership probabilities for each of the considered populations to belong at each of the two sPC1-identified groups (white = South-Eastern Mediterranean; black = North-Western Mediterranean). Population codes as in Table S1. Fisher exact tests were carried out among groups of populations in order to identify significantly over- or under-represented HGs in any of the geographic areas analysed, against a background of all the other Mediterranean populations (Table S6). Haplogroup G-M201 appears significantly over-represented in the SSI genetic pool. Haplogroup R-M269, has been found significantly over-represented in Western-Mediterranean populations (IBE, GER and NCI), and under-represented in the South-Eastern Mediterranean ones (BALK, LEV and NAFR). By contrast, haplogroup J-M304(xM172) is significantly over-represented in the non-European Mediterranean shore (LEV and NAFR), being instead under-represented in European Mediterranean populations. In order to investigate further, we then performed a set of Bonferroni-corrected Chi-square tests by comparing frequencies of single lineages in SSI with those of each reference Mediterranean population group, this time exploiting the highest Y-SNP level of resolution available for each pairwise populations comparison (and considering only those lineages with absolute frequency of at least 10 individuals in SSI). Being aware that migration processes cannot be linked only with single specific haplogroups, it is however known that signals of migration should be more easily detected in more highly differentiated lineages [44]. Different haplogroups have shown significantly higher frequency in specific comparison groups than in SSI: R1b-sublineages in the western European samples (R-U152 for North-Central Italy, P-value<0.001; R-P312 for Iberian Peninsula, P-value<0.001; and R-U106 for German region, P-value<0.001), R-M17 in the Balkan Peninsula and Germany (both P-values<0.05), and J1-M267 in both Levant and North-Africa (both P-values<0.001).  Table 1. Age estimates (in YBP) of STR and HVS variation for the most frequent haplogroups in Sicily and Southern Italy. As for TMRCA estimates, STR variation within the most frequent haplogroups of SSI suggests that most of them (with the exception of haplogroup G2a-P15: 9339±3302 YBP) date back to relatively recent times (Table 1), in some cases falling into time periods compatible with specific documented historical events occurred in SSI. Despite the fact that these time estimates must be taken with caution, as they might be affected by the choice of both STRs markers and their mutation rates, overall our results agree in suggesting that most of the Y-chromosomal diversity in modern day Southern Italians originated during late Neolithic and Post-Neolithic times (∼2,300 YBP for E-V13; from ∼3,200 to ∼3,700 YBP for J sub-lineages; ∼4,300 YBP for R-M17 and R-P312; and ∼2,000 YBP for R-U106 and R-U152). |
|
|
|
Post by Admin on Sept 18, 2019 17:39:24 GMT
Mitochondrial DNA perspective The maternal genetic ancestry of SSI population was explored by successfully typing both coding region SNPs and HVSI-HVSII sequences in 313 out of the 326 samples. Overall, the polymorphic sites observed in the D-loop and coding region allowed assignment of subjects to 40 mtDNA HGs (including sub-lineages), whose frequencies for both the whole dataset as well as for each of the 8 sampling points are reported in Table S2. In order to ensure the easiest access to the data [45], mtDNA sequences were deposited in the GenBank nucleotide database, under accession numbers KJ522492-KJ522611. The observed mtDNA HGs distribution reflects the typical maternal variability pattern documented for Mediterranean Europe. In fact, most of the individuals belong to super-haplogroup H, that on the whole accounts for the 38% of the total mtDNA lineages detected in our dataset. Within H, H1 represents the most frequent sub-lineage (10.9%), followed by H5 (3.2%) and H3 (2.6%). Noteworthy is also haplogroup HV, that has been found at relatively high frequencies (4.8%). Most of the remaining samples belong to haplogroups U5, K1, J1, J2, T1, T2, thus confirming prevalent European and Middle-Eastern genetic ancestries. MtDNA haplotypes of African origin are instead represented by few haplogroups at low frequencies, namely M1 (1.3%), U6a (0.6%) and L3 (0.6%). Within-population diversity indices reveal that, in the context of our dataset, Sicily (and particularly Western Sicily) shows slightly lower diversity values than Southern Italy (Table S5). Nevertheless, the diversity parameters observed for all the 8 populations analysed as well as for the whole dataset, fall within the range of values commonly reported in literature for both Italian and Southern European populations [11]. Similarly to Y-chromosome, mtDNA does not reveal any kind of population sub-structure both within Sicily (East vs. West Sicily) as well as between Sicily and Southern Italy, neither considering haplogroups nor haplotypes (sequences). AMOVA results show low and non-significant FCT values when population samples were grouped according to geography (Table S4). Analogously, Fisher exact tests reveal no significantly different HG composition in any of the geographic regions considered (South Italy vs, Sicily, P-value: 0.5019; East Sicily vs. West Sicily, P-value: 0.0698). In the same way, both AMOVA (variation among groups 0.52%, P-value 0.082) and Fst (P-value 0.076) based on HG frequencies show the absence of significant genetic differentiation along the east-west axis of Sicily.  Figure 3. Spatial Principal Component Analysis (sPCA) based on mtDNA haplogroups frequencies. The first two global components sPC1 (a) and sPC2 (b) are depicted. Positive values are represented by black squares; negative values are represented by white squares; the size of the square is proportional to the absolute value of sPC scores. The mtDNA HGs geographic distribution within the Mediterranean domain was investigated by comparing our sample with 26 Euro-Mediterranean, Levantine and North-African populations selected from the literature (Table S1). A Mantel test shows a low correlation between geographic and genetic distances (observed value = 0.279, P-value = 0.016). In order to further explore the relationships between geography and mtDNA genetic variability, we performed a sPCA (using HG frequencies). The highest eigenvalue obtained is the most positive one (sPC1) associated with the presence of a global structure. As previously emerged for Y-chromosome, sPC1 plot reveals a North-West/South-East (NW-SE) distribution of mtDNA genetic variation (Figure 3a). Nearly all of the Mediterranean populations (with some exceptions, i.e. AG, TV, BUR) appear indeed distributed along a longitudinal transect running from North African and Near Eastern countries (large white squares) to the Iberian Peninsula (large black squares), with the bulk of the South-Eastern European populations (including Balkans and Italy) roughly occupying an intermediate position therein (see also Figure S2b). Among them, Sicily and Southern-Italy appear linked to the South-Eastern Mediterranean coast. When the reliability of this sPC1-identified structure has been tested by means of AMOVA, the proportion of genetic variation between groups (FCT) results lower than in the case of Y-chromosome (2.45%) but still significant (P-value<0.001). The second sPC (Figure 3b) highlights the position of Italy within the Mediterranean context and particularly of its South-Eastern part (large white squares). However, when tested with AMOVA, the proportion of variation between groups (FCT) explained by sPC2 revealed to be not significant (0.48%, P-value = 0.212). On the whole, the lack of statistical support for the global structure observed in the mtDNA sPCA (Gtest: obs = 0.165, P-value = 0.065), suggests a higher homogeneity in Mediterranean genetic variability for maternal than paternal genetic pools. Nevertheless, both uniparental markers show a similar NW-SE distribution pattern of genetic variation. Fisher exact tests were applied to determine if differences in HG frequencies among population groups were statistically significant (Table S6). As expected, haplogroup H is found to be over-represented in Euro-Mediterranean populations and under-represented in North-African ones, while the opposite has been observed for haplogroup L. Haplogroup K is over-represented in Levantine populations, and haplogroup M in North-Africa. However, when the deepest level of HG resolution has been exploited for single pairwise comparisons between SSI and Mediterranean reference populations, we do not found any HG whose frequency is significantly higher than in our dataset. The only exception is a slightly significant (P-value: 0.045) over-representation of H1 haplotypes in the Iberian Peninsula. Differently from Y-chromosome results, TMRCA estimates for the most frequent mtDNA haplogroups of Sicily and Southern Italy (Table 1) date back to pre-Neolithic times and could be mainly classified in lineages pre-dating the Last Glacial Maximum - LGM (∼32,200 YBP for HV; ∼31,100 YBP for J2; ∼28,900 and ∼28,600 YBP for T1 and T2; ∼27,300 for U5; and ∼25,000 YBP for J1) or dating immediately after it (∼16,700 YBP for H5 and ∼15,700 YBP for H1). Comparative analysis of maternal and paternal genetic pools The admixture-like plot represented in Figure 4 summarizes the genetic relationships between SSI and the chosen Mediterranean populations by directly comparing Y-chromosome and mtDNA genetic results.  Figure 4. Admixture-like barplots for Y-chromosome (a) and mtDNA (b). The barplots represent DAPC-based posterior membership probabilities for each of the considered populations and for each inferred cluster (mclust algorithm). The affiliation of each population to a given cluster and its corresponding colour code are represented by letters (within coloured squares) on the top of each bar. Labels: NAFR: North-Africa, LEV: Levant, BALK: Balkans, SSI: Sicily and South-Italy, NCI: North-Central Italy, IBE: Iberian Peninsula, GER: Germany. From a Y-chromosome point of view, SSI form a fairly coherent group with the Levantine and the Balkan populations (cluster 2), despite showing some minor contribution (black component) also from the North-Western Mediterranean group (cluster 3). From a mtDNA point of view, our results show the differentiation between European and non-European Mediterranean populations, with North Africa and the Levant clustering in separate and different groups (1 and 2). However – and differently from the other European populations – SSI shows a noteworthy contribution (grey component) from the Levantine cluster. Both genetic systems reveal a negligible contribution from North Africa (white component). The extent of different contributions to the current SSI genetic variation was further assessed by means of an admixture analysis performed (on HG-frequencies) with the coalescent-based mY estimator implemented in the software Admix 2.0 [39]-[40]. We used a tri-hybrid admixture model, considering as source populations North-Western Italy, the Balkans and the Levant (see Materials and Methods for more details). While keeping in mind that selection of parental populations can potentially misrepresent the real estimate of admixture proportions [41]–[43], our admixture rates (Figure S3) are however quite consistent with the above-mentioned results (despite the high standard errors values). Y-chromosome admixture proportions to the current SSI genetic pool indeed confirm an high paternal contribution from the South-Eastern Mediterranean populations, and particularly from the Balkan Peninsula (∼60%), whereas about 25% of SSI Y-chromosomes can be traced back to North-Western European group. Analogously, although the present-day SSI mtDNA genetic pool is largely shared with the other South-Eastern European populations of the Mediterranean Basin (respectively Balkan and Italian Peninsulas), a remarkable proportion of maternal ancestry (especially if compared with its paternal counterpart) derives from the Levant. |
|
|
|
Post by Admin on Sept 18, 2019 20:21:09 GMT
Discussion and Conclusions Sicily and Southern Italy have long represented a natural hub for the expansion of human genes and cultures within the Mediterranean Basin [1]. Accordingly, the genetic pool of current populations inhabiting this area can be interpreted as the result of complex interplays and superimpositions between different prehistoric and more recent demographic events, ranging from the Neolithic expansion and the proto-historic Greek and Phoenician colonisations, up to the post-Roman invasions by Byzantines, Arabs and Normans. The real demographic impacts of these settlements on the population structure remain still largely uncertain based on the study of material culture and the available historical sources, and different hypotheses about the relative contributions of these events to the current gene pool composition have been proposed from a genetic point of view [7]–[9]. As a contribution to the human history of such a key area of the Mediterranean we surveyed, by means of a comprehensive evaluation of both maternal and paternal genetic landscapes, the genetic variability of a wide number of populations settled in a broad transect encompassing Sicily and Southern Italy (Figure S1). Previous reconstructions of the genetic structure of Sicily [7]–[9] focused their attention mainly on two points in the attempt to clarify its genetic history: a) the presence or absence of internal genetic differentiation along an east-west axis, and b) the extent of the genetic relationship with other populations of the Mediterranean Basin.  Population structure and genetic history of Sicily and Southern-Italy In contrast with previous investigations on the distribution pattern of genetic variation in Sicily [7]–[8], our results point to a substantially homogeneous composition of maternal and paternal genetic pools both within Sicily (East vs. West) as well as between Sicily and Southern Italy (Table S4). The absence of significant differences in the distribution of HG frequencies along the east-west axis of the island, as observed not only among our Sicilian populations, but also when including the samples from Di Gaetano et al. (2009) [8], provides further support to these conclusions. The comparison of the whole SSI dataset with a subset based on founder surnames, moreover suggests that the observed homogeneity in Y-chromosome composition is not the result of recent events (e.g. increased population mobility related to the social and economic changes of the 19th and 20th centuries); on the contrary it has been preserved at least since the initial founding and spreading of surnames in Italy. In addition, and consistently with the complex history of migration pathways and cultural exchanges characterizing the peopling history of the area, high levels of Y-chromosome and mtDNA genetic variability at both SNP and haplotype (STRs or sequence) data, have been observed in all the SSI populations here examined (Table S5). Altogether, the high levels of within-population variability and the lack of significant genetic sub-structures fit well with the historic role of Sicily and Southern Italy as a major migration crossroad within the Mediterranean Basin. Anyway, differential contributions from the considered Euro-Mediterranean areas were observed. For instance, if the Near East, the Balkans, and – at a lesser extent – North-Western Italy probably had a relevant role in the genetic make-up of SSI, Northern African contributions seem to be almost negligible. As for the Iberian Peninsula, at present its specific genetic contribution cannot be distinguished from that of North-Western Italy, given their observed genetic similarity. These multiple migration events have probably favoured the reduction of genetic differentiation across the region, by increasing the rates of gene flows between different ethnic groups and in some cases mixing up the different genetic strata. Interestingly, the presence of massive migratory phenomena not necessarily yields genetic homogeneity in a given region. For instance, recent studies [46]–[47] showed how ethno-linguistic minorities from Sicily and Southern Italy - such as the Albanian-speaking Arbereshe - may conserve a significant genetic diversification from the rest of the population. In general, such features are more easily observed in isolated populations, thanks to their reduced population size and their cultural distinctiveness, if compared to open populations. The patterns of genetic variability observed in our SSI sample are in agreement with the general statement that Southern European populations tend to show higher levels of genetic diversity when compared with those located at more northern latitudes [48] by virtue of the several past demographic events that affected their genetic composition over time. Additionally to the postglacial re-expansion and the demic diffusion of agriculture from Near East, also more recent events (e.g. gene flows from North Africa [48]) have been recently advocated as other possible explanations for the increased genetic diversity in the Southern European populations. Among the several historical occupations of Sicily and Southern Italy, the Pre-Roman colonisation by Greeks and Phoenicians as well as the subsequent invasions from North Africa (including the Muslim conquest, that, at least in part, was conducted by Berber forces) have been previously suggested as putative contributors to the gene pool of current Sicilian population (at least from a male perspective [8]). At this respect, the distribution of Y-chromosome haplogroup E-M81 is widely associated in literature with recent gene flows from North-Africa [49]. Besides the low frequency (1.5%) of E-M81 lineages in general observed in our SSI dataset, the typical Maghrebin core haplotype 13-14-30-24-9-11-13 [8] has been found in only two out of the five E-M81 individuals. These results, along with the negligible contribution from North-African populations revealed by the admixture-like plot analysis, suggest only a marginal impact of trans-Mediterranean gene flows on the current SSI genetic pool. Together with the Berber E-M81, the occurrence of the Near-Eastern J1-M267 in Southern-European populations has been linked to population movements from the Near East through North-Africa, and particularly as a marker of the Islamic expansion over Southern-Europe (started approximately in the 8th century AD and lasted for more than 500 years). Fisher exact tests based on HGs frequencies have revealed the presence of haplogroup J1-M267 at significantly higher frequencies in both North-Africa and the Levant than in Sicily and Southern Italy (both P-values<0.001). However, the estimated age for Sicilian and Southern-Italian J1 haplotypes refers to the end of the Bronze Age (3261±1345 YBP), thus suggesting more ancient contributions from the East. Nevertheless, our time estimate does not necessarily coincide with the time of arrival of J1 in SSI; in fact a pre-existing differentiation could potentially backdate the time estimate here obtained. By the collapse of the Late Bronze Age societies (approximately 3200 YBP), the Mediterranean Basin underwent different waves of invasion, particularly by the Greeks of the Aegean Sea and, to a lower extent, by Levantine (Phoenicians) groups [50]. Both of them established a set of different colonies along the Mediterranean coasts of Southern Europe and North Africa. The Phoenician colony of Carthage (present-day Tunisia), given its geographic proximity to Sicily, may have played an important role in the colonization of this region. Previous Y-chromosome genetic studies on the Phoenician colonization demonstrated that haplogroup J2 in general, and six haplotypes in particular (PCS1+ through PCS6+), may potentially have represented lineages linked with the spread of the Phoenicians (“Phoenician Colonization Signal”) into the Mediterranean [51]. At this respect, it is worth noting the presence of 4 PCS+ haplotypes (namely PCS1+, PCS2+, PCS4+, PCS5+; [51]) in 9 samples of our Sicilian and Southern Italian dataset, particularly belonging to haplogroups J1-M267 (n = 2), J2-M410* (n = 1), J2-M67 (n = 5), and J2-M12 (n = 2). However, sub-lineages of haplogroup J2 have been also associated with the Neolithic colonization of mainland Greece, Crete and Southern Italy [52], and our TMRCA estimates for J2-subhaplogroups (ranging from 3271±1157 YBP to 3767±1332 YBP) cannot exclude an earlier arrival of at least some of the J2 chromosomes in Sicily and Southern-Italy during Neolithic times.  On the other hand, Y -chromosome lineage E-V13 is thought to have originated in southern Balkans [53]–[54] and then to have spread in Sicily at high frequencies with the Greek colonization of the island [8]. The E-V13 core haplotype 13-13-30-24-10-11-13 (DYS19-DYS389I-DYS389II-DYS390-DYS391-DYS392-DYS393), which define the southern Balkan Modal Haplotype and reaches frequencies of ∼12% in continental Greece [52], has been found in 10 out of the 31 E-V13 samples of Sicily and Southern Italy. This result, along with the high frequency of E-V13 lineages generally observed in our dataset (the second most frequent haplogroup after G2a), confirms the presence of gene flows into Sicily from the Balkans as previously observed by Di Gaetano et al. (2009) [8]. Accordingly, our TMRCA estimate for E-V13 (2354±832 YBP) agrees with the results previously reported in literature for the Sicilian population (2380 YBP, [8]). Altogether, these results do not exclude the possible introduction of some of these Y-lineages with migration processes originated in the Balkans and particularly associated with the Greek colonisation of Southern Italy. Y-chromosome haplogroup G2a-P15 turn out to be of particular interest in the paternal genetic make-up of Sicily and Southern Italy. Its older age estimate (9339±3302 YBP) – if compared to those of other haplogroups – along with its significantly over-represented frequency in SSI, are consistent with the hypothesis recently suggested by Boattini et al. (2013) [12] according to whom this lineage could be a possible candidate for a pre-Neolithic ancestry in Italy. However the CIs of our time estimate cannot exclude alternative hypotheses such as a diffusion of its major sub-clades during Neolithic and Post-Neolithic times, as recently discussed by Rootsi et al. 2012 [55]. Contrarily to Y-chromosome results, age estimates for mtDNA haplogroups suggest that most of the maternal diversity of the current Sicilian and Southern Italian population is composed by lineages present in Europe as early as the LGM (Table 1). The Late Glacial and Postglacial re-occupation of Europe from refugial areas located in the Mediterranean Peninsulas, has played a major role in shaping the gene pool of modern Europeans [56] and some of the differences in genetic diversity of current European populations have been attributed also to this process [48]. Consistently, the geographic distribution and ages of some mtDNA haplogroups, such as V, H1 and H3, have been associated to events of postglacial re-colonisation from Southern European glacial refugia, and particularly from the Franco-Cantabrian area [57]–[60]. Further evidences of post-glacial resettlement from Southern refugia have been recently suggested also for the mtDNA haplogroup H5 (the third most common European H-sublineage after H1 and H3), if considering its higher occurrence in southern European populations (particularly Italy) and its evolutionary age ranging approximately between 11,500 and 16,000 YBP [61]. Together with the Iberian and Balkan peninsulas, also Italy and particularly SSI might have played an important role during the post-glacial re-expansion, as widely attested by several animal and plant species [62]–[68]. As in the case of Iberia and the Balkans, the presence of numerous Epigravettian sites suggests that Italy could have acted as such also for humans [69], despite the fact that strong genetic evidences are still missing (except for mtDNA haplogroup U5b3 [70]). Haplogroups H1 and H5 appeared to represent the most frequent H-sublineages in SSI, and their age estimates (Table 1) are consistent with post-glacial time periods, as previously observed for both Southern Italy [11] and the entire Peninsula [12]. Nevertheless, a significant (P-value 0.045) over-representation of H1 haplotypes and an older age (17295±5119 YBP) has been obtained for the Iberian population (as represented by the considered reference samples) than in our SSI datatset, thus suggesting, at least for H1,a post-glacial re-expansion presumptively originated in the Franco-Cantabrian area. Interestingly, mtDNA haplogroup HV confirmed to be the most ancient lineage in Sicily and Southern Italy, predating the LGM (32242±12595 YBP) and thus representing a possible candidate for the Palaeolithic ancestry of Southern Italy, even though possible post-LGM expansions of its major sub-branches should be taken into account as potentially affecting the time estimates here obtained. Further analyses, involving the complete sequencing of mtDNA genomes and the analysis of ancient DNA samples, are therefore needed in order to more deeply address this point and to confirm the relevance of this haplogroup in the first peopling of Sicily by moderns humans, as recently suggested by some Palaeogenetic researches [5]. Sarno S, Boattini A, Carta M, Ferri G, Alù M, Yao DY, et al. (2014) An Ancient Mediterranean Melting Pot: Investigating the Uniparental Genetic Structure and Population History of Sicily and Southern Italy. PLoS ONE 9(4): e96074. |
|
|
|
Post by Admin on Sept 19, 2019 18:03:44 GMT
Recently, genome-wide analyses brought new attention on different aspects of the genetic history of Greece and the Balkans11, 12, 33, especially since ancient paleogenomic data became available for Anatolia and Northern Greece5, 8, 34. On the other hand, while aDNA data from Southern Italy and Sicily are still limited, genome-wide analyses of modern populations from these areas mainly consisted to wide-range surveys without specific fine-scale insights18, 35, 36. In this study, we genotyped 511 samples belonging to 23 populations from Sicily, Southern Italy, Albania and Greece, as well as from Italian Arbereshe and Greek-speaking ethno-linguistic groups (Fig. 1, Supplementary Table S1, Supplementary Information). By comparing our data with a large collection of modern and ancient populations from Europe and the Mediterranean, we aim to address the following questions: (1) How does Southern Italy fit within the broader context of the Mediterranean genetic landscape and with respect to Southern Balkan and Greek populations? (2) What can we suggest about the peopling of the area in terms of ancient admixture layers and more recent historical contributions? (3) Is there any evidence of genetic links between the Arbereshe and Greek-speaking ethno-linguistic minorities of Southern Italy and their putative populations of origin? Could our data provide additional insights into their demographic history as recent “cultural islands”? 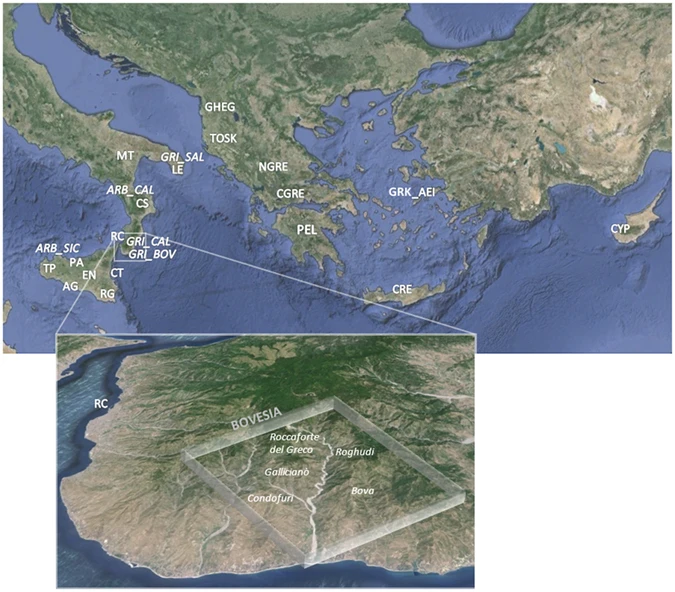 Figure 1 Population structure and admixture To provide a first overview of the geographic and temporal relationships between our newly analysed populations and the Euro-Mediterranean genetic landscape, we assembled an extended dataset consisting of 1,469 individuals from 68 modern populations, together with 263 ancient samples (Supplementary Table S2, Supplementary Table S3), and we ran PCA and ADMIXTURE analyses (Fig. 2, Supplementary Fig. S1, Supplementary Fig. S2, Supplementary Fig. S3, Supplementary Information). 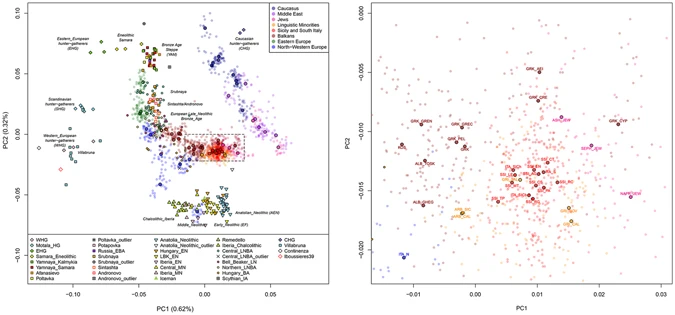 Figure 2 Principal component analysis performed on the extended comparison dataset with ancient samples projected onto the first two PCs. (a) Scatterplot of the first and second PCs for 1,469 modern samples from 68 Euro-Mediterranean populations and 263 projected ancient samples. Ancient individuals are labelled and symbol-coded according to their associated culture, as reported in the legend at the bottom of the plot and detailed in Supplementary Table S3. Modern individuals and median population coordinates (enlarged black-bordered circles) are colour-coded based on their geographic or ethnic affiliation, as in the legend at the top-right and in Supplementary Table S2. (b) A magnification of the plot details the position of the newly analysed Southern Italian and Southern Balkan populations within the observed large-scale genetic diversity. Modern Southern Italian and Southern Balkan populations are located at the centre of the PCA plot (Fig. 2, Supplementary Fig. S1), forming an almost uninterrupted bridge between the two parallel clines of distribution where most of the other modern populations are found, one stretching along the East-West axis of Europe and the other from the Near East to the Caucasus, respectively (see also Supplementary Information). In particular, Sicily and Southern Italy (SSI) appear as belonging to a wide and homogeneous genetic domain, which is shared by large portions of the present-day South-Eastern Euro-Mediterranean area, extending from Sicily to Cyprus, through Crete, Aegean-Dodecanese and Anatolian Greek Islands. We will refer to this domain as ‘Mediterranean genetic continuum’. On the other hand, the continental part of Greece, including Peloponnesus, appears as slightly differentiated, by clustering with the other Southern Balkan populations of Albania and Kosovo. Finally, North-Central Balkan groups (Southern Slavic-speakers and Romanians) show affinity to Eastern Europeans (Fig. 2, Supplementary Fig. S1, Supplementary Information). Admixture results further show our newly analysed populations as a blend of the major ancestry genetic components detected in the broader Mediterranean region, namely the European-like, Caucasian-like, Sardinian-like and Near Eastern-like ones (Supplementary Fig. S2a, Supplementary Information). Importantly, three of them find empirical correspondence (see Supplementary Information for more details) to ancient population ancestries, represented respectively by European Hunter Gatherers, Caucasus Hunter Gatherers and Early Neolithic farmers (Supplementary Fig. S3). All populations from Southern Italy (SSI), Greece (both mainland and insular) and Southern Balkans share a predominant Sardinian (Neolithic-like) genetic component which accounts for more than half of their ancestry. This is followed by a relevant Caucasian-like ancestry, which is present at around 24% in all our population samples (Supplementary Fig. S2b). The other two major components instead show opposite patterns. The Near Eastern-like ancestry is more frequent in SSI and the Greek-speaking islands (i.e. the ‘Mediterranean continuum’), whereas increasing frequencies of the European-like component are observed in Albanians and mainland Greeks as well as in the rest of the Balkan Peninsula (Supplementary Fig. S2b). Interestingly, Grecani of Calabria (GRI_BOV and GRI_CAL) and Cypriots share lower frequencies of the European-like ancestry (2.5% and 0.5%, respectively) compared to the other surrounding populations (Southern Italy: ~8%; Continental Greece and Albanians: ~15%). |
|










