|
|
Post by Admin on Nov 14, 2019 18:43:56 GMT
Autosomal SNP-s We have predicted eye, hair and skin color phenotypes from 25 HirisPlex SNP-s, also suitable to predict non-European ancestry14, as summarized in Figs 3 and 4. Samples from different archaeological cultures and cemeteries showed a remarkable pattern of phenotypic distribution. All Hun and Avar age samples had inherently dark eye/hair colors, DK/701 being the only exception (Fig. 3). Moreover 6/14 Avar age samples were characterized with >0,7 black hair; >0,99 brown eye p-values, inferring 86,5% probability of non-European biogeographic ancestry14 in agreement with their anthropological, archaeological and historical evaluation. In contrast the Conquerors showed a wide variety of phenotypes clustered by cemeteries (Fig. 4). All individuals from the Sárrétudvari (SH), Magyarhomorog (MH) and majority from the Kenézlő (KEF) graveyards displayed European phenotypic patterns; blue eye and/or light hair with pale skin. In the three Karos cemeteries darker eye/hair colors predominated, 4/20 individuals having p-values consistent with non-European origin, nevertheless 5/20 individuals had light hair color indicating a rather mixed origin of this population, concurrent with their mtDNA and Y chromosomal Hg composition. 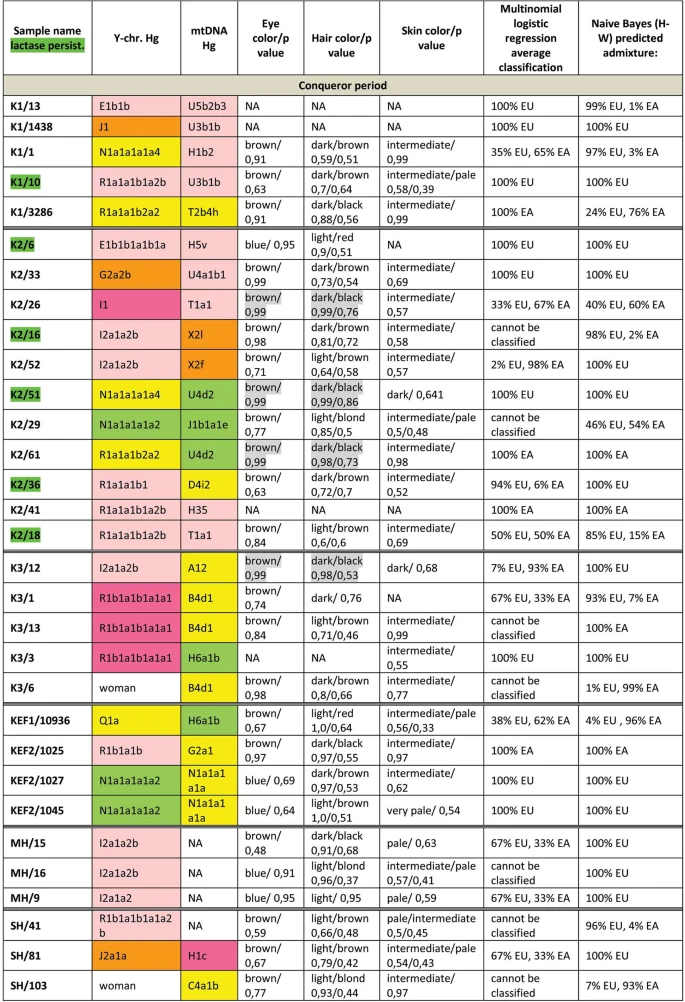 Figure 4 We have also determined 34 AIMs, which can inform about the likely geographic origin of individuals, and used the Snipper App suite version 2.5 portal16 to assign biogeographic ancestry. We predicted ancestry with both the naive Bayesian classifier and multinomial logistic regression (MLR) algorithms, as these make different assumptions about genetic equilibrium28, and listed the results on Figs 3 and 4. The AIM-s results fairly matched and complemented phenotypic information. All Hun age individuals revealed admixture derived from European and East Asian ancestors, while 8/15 Avar age individuals showed predominantly East Asian origin with both methods, 4 individuals were definitely European, while two showed evidence of admixture. The KFP/31 sample gave contradicting results due to low coverage. Conqueror samples from the Magyarhomorog (MH) and Sárrétudvari (SH) cemeteries showed mostly European ancestry in agreement with their phenotypes and Y Hg-s, though MLR detected a significant east Asian ancestry component and the SH/103 women was classified east Asian despite her blond hair. The Karos (K) and Kenézlő (KEF) populations were profoundly admixed, comprising individuals of purely East Asian, European and mixed origin in nearly identical proportions, again in agreement with results obtained from uniparental and phenotypic markers. The determined variable autosomal loci are also suitable to exclude possible direct (parent-child, sibling) genetic relatedness29, thus we compared the autosomal genotypes of all individuals sharing either maternal or paternal Hg-s. Direct kinship could not be excluded between the MH/9 and MH/16 individuals with identical phenotypes, suggesting that MH/9 probably also belongs to Hg I2a1a2b-L621, despite its uncovered L621 marker. KEF2per1027 and KEF2per1045 were probably brothers as they had identical mitogenomes, Y Hg-s and blue eye color besides sharing autosomal alleles. The same applies to K3/1 and K3/13 individuals who were probably also brothers. We could not exclude possible direct paternal relationship between K2/36, K2/18 and K2/41 but the first two samples had unsatisfactory coverage to make a strong statement. We have tested two SNP-s (rs4988235 and rs182549) associated with the adult lactase persistence phenotype in Europe15. Individuals carrying derived alleles in these loci are able to digest lactose in dairy products during adulthood without symptoms of lactose intolerance. Allele frequency of the persistence genotype varies throughout Eurasia30, reaching above 90% at some parts of Northwestern Europe, around 80% in present day Hungary, but drops below 30% in Central Asia and even lower in East Asia. We detected the derived persistence allele in all of the studied groups (Figs 3 and 4, Supplementary Table S2); 1/3 of the Hun period individuals, 2/14 of the Avar period individuals, and 6/31 of the Conqueror period individuals carried persistence alleles. It is remarkable that the persistence genotype seems to be associated with European origin, as all of the carriers were predicted to have predominantly European ancestors. This is in agreement with a previous study31, which found that all of the 11% Conqueror samples with persistence genotype carried European mtDNA Hg H. In addition all carriers were heterozygous, 6 of them for both SNP-s, but three of them carried just the rs182549 (-22018 G > A) derived allele suggesting previous admixture with non-carriers, possibly derived from East Eurasia. |
|
|
|
Post by Admin on Nov 15, 2019 18:12:37 GMT
Population genetic analysis The studied Conqueror group very likely represent real populations, as 24 of the 29 samples came from 4 nearby cemeteries (Karos 1,2,3 and Kenézlő) with identical archaeological and anthropological groupings and the studied Magyarhomorog individuals were also categorized as belonging to the same early Conqueror elite. The Avar group was assembled from several different cemeteries of a wider timespan, thus they cannot represent the Avar period population of the Carpathian Basin, however their relatively homogenous Hg distribution indicate that the Avar elite embodied the same east Eurasian sub-population throughout their reign, so it appeared to be meaningful to include them in the analysis. In order to find the most similar populations to our studied ones, we compared the Hg distribution of the Avar and Conqueror elite to that of 78 modern Eurasian populations (Supplementary Table S4) and represented their relations on MDS plot displayed on Fig. 5. Only one individual was included in the analysis from first degree relatives. In order to increase resolution, we separately calculated the frequencies of each sub-Hg-s which are present in our samples, while frequencies of all other subclades were combined as listed in Supplementary Table S4. Similar Hg distributions are mapped into neighboring positions on the MDS plot, which clearly separates populations according to their geographic locations. Along the x axis east Eurasian populations map to the right while Europeans are compressed at the left. Along the y axis Siberian populations are sequestered at the top, while East Asian ones at the bottom of the graph. The Avars are obviously mapped to the Siberian side with smallest weighted Euclidean distances from Koryaks (KRY), Teleuts (TEL), Khantys (KHT), Komis, (KOM) and Dolgans (DLG). The Conquerors are positioned between eastern Europeans, Central Asians and Siberians but their exact relations are hard to make out because of the crowding at the European side. 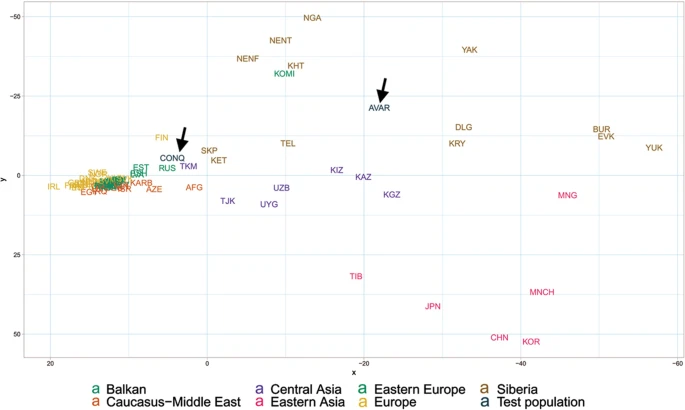 Figure 5 In order to better discern the position of the Conquerors we redraw the MDS plot without the eastern Asian and Siberian populations (Fig. 6). Though having removed subset of the data MDS imaging in two-dimensions inevitably rearranged the remaining components, but the general organization of the population clusters remained unchanged: Western-Northern Europeans map to the left, Central-eastern European and Balkan people around the middle, Central Asians to the top right, and Caucasus-Middle East populations to bottom right. Conquerors remained between Central Asians and eastern Europeans, with smallest weighted Euclidean distances from Bashkirs (BSH), Hungarians (HUN), Tajiks (TJK), Estonians (EST), Kazakhs (KAZ), Uzbeks (UZB) and Slovaks (SVK). 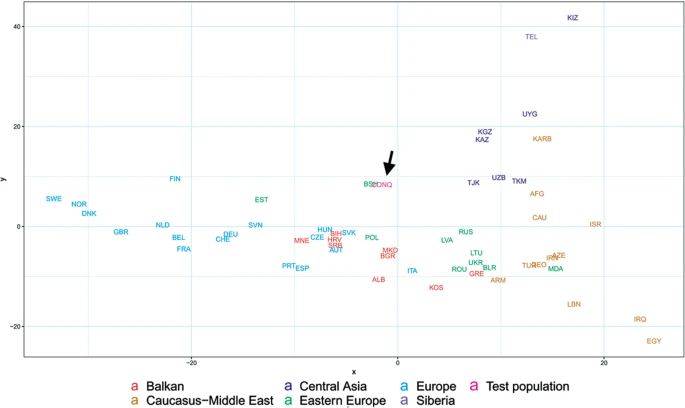 Figure 6 The considerable frequency of Hg N1a in Conquerors and especially in Avars facilitates another analysis in which the frequency distribution of their N1a subbranches can be compared to that of all Eurasian populations carrying this Hg, which is described in22. The N1a database of22 contains several relevant Eurasian populations missing from Supplementary Table S3, moreover the Y dataset of Supplementary Table S4 has a low resolution of N1a subclades, thus this analysis is expected to provide additional information. We combined the N1a dataset published in22 with our data as presented in Supplementary Table S5 and performed Principal Component Analysis (PCA) as shown on Fig. 7. 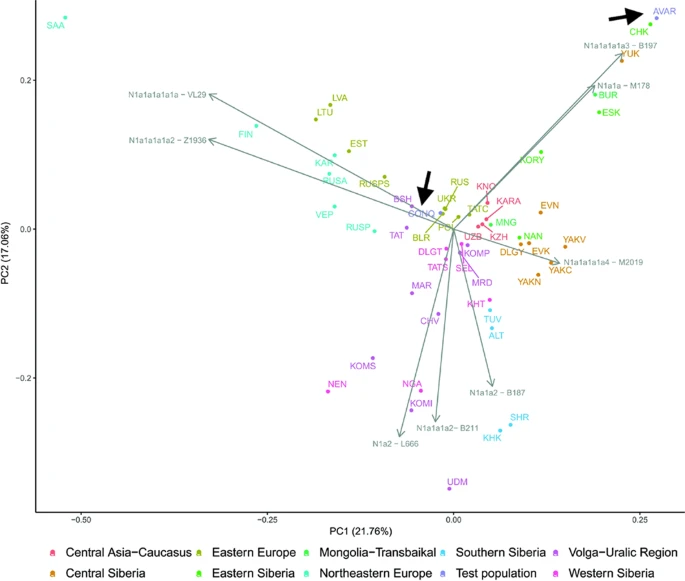 Figure 7 PCA also separates populations according to their geographical positions, as identical colors appear in the same segments of the graph. The Avars map to the top right corner, close to eastern Siberian Chukchis (CHK), Yukaghirs (YUK), Eskimos (ESK) and Transbaikalian Buryats (BUR). The Conquerors (CONQ) map to the middle of the graph, very close to eastern European Belarusians (BLR), Ukrainians (UKR), Russians (RUS) and Polish (POL), not far from Volga-Uralic region Bashkirs (BSH), Volga Tatars (TAT), Komi Permians (KOMP) and Mordvins (MRD), also surrounded by Central Asian groups; Karanogays (KNO), Karakalpaks (KARA), Uzbeks (UZB) as well as Crimean Tatars (TATC), Mongols (MON), Selkups (SEL) and Dolgans from Taymyr (DLGT) |
|
|
|
Post by Admin on Nov 16, 2019 6:31:45 GMT
Discussion
The origin and composition of the Conqueror paternal lineages fairly mirrors that of their maternal ones10; 20,7% of the Y-Hg-s originated from East Eurasia, this value is 30,4% for mtDNA; proportion of west Eurasian paternal lineages is 69% compared to 58,8% for mtDNA; while proportion of lineages with north-western European and Caucasus-Middle East origin are nearly the same affirming that both males and females of similar origin migrated together. Both MDS analysis of the entire Conqueror Y chromosome pool and PCA of their N1a lineages indicates that their admixture sources are found among Central Asians and eastern European Pontic Steppe groups, a finding comparable to what had been described for maternal lineages10. Composition of the Conqueror paternal lineages is very similar to that of Baskhirs, while their maternal composition was found most similar to Volga Tatars10. These modern populations are located next to each other, have similar prehistory32 and genetic structure derived from the same admixture sources detected in the Conquerors. Moreover it must be noted that Bashkirs were not represented in the mitogenome database while Volga Tatars are missing from our Y-chromosome database due to lack of data, but their N1a distribution is quite similar (Fig. 7), thus mtDNA results are in accord with Y-chromosomal ones.
The Conqueror-Bashkir relations are also supported by historical sources, as early Hungarians of the Carpathian Basin were reported to be identical to Baskhirs by Arabic historians like al-Masudi, al-Qazwini, al-Balhi, al-Istahri and Abu Hamid al-Garnati33, latter visited both groups at the same time around 1150 AD and used the term Bashgird to refer to the Hungarians in the Carpathian Basin. In addition parallels were found between several Conqueror and Bashkir tribe names and Bashkiria has been identified with Magna Hungaria, the motherland of Conquerors34.
We identified potential relatives within Conqueror cemeteries but not between them. The uniform paternal lineages of the small Karos3 (19 graves) and Magyarhomorog (17 graves) cemeteries approve patrilineal organization of these communities. The identical I2a1a2b Hg-s of Magyarhomorog individuals appears to be frequent among high-ranking Conquerors, as the most distinguished graves in the Karos2 and 3 cemeteries also belong to this lineage. The Karos2 and Karos3 leaders were brothers with identical mitogenomes10 and Y-chromosomal STR profiles (Fóthi unpublished). The Sárrétudvari commoner cemetery seems distinct from the others, containing other sorts of European Hg-s. Available Y-chromosomal and mtDNA data10 from this cemetery suggest that common people of the 10th century rather represented resident population than newcomers. The great diversity of Y Hg-s, mtDNA Hg-s, phenotypes and predicted biogeographic classifications of the Conquerors indicate that they were relatively recently associated from very diverse populations.
In contrast the studied Avar military leader group had a much more uniform origin. The Avar group carried predominantly East Eurasian lineages in accordance with their known Inner Asian origin inferred from archaeological and anthropological parallels as well as historical sources. However the unanticipated prevalence of their Siberian N1a Hg-s, sheds new light on their prehistory. Accepting their presumed Rouran origin would implicate a ruling class with Siberian ancestry in Inner Asia before Turkic take-over. The surprisingly high frequency of N1a1a1a1a3 Hg reveals that ancestors of contemporary eastern Siberians and Buryats could give a considerable part the Rouran and Avar elite, nevertheless a larger sample size from more Avar cemeteries are needed to clarify their exact composition.
The genetic profile of the Avar and Conqueror leader groups seems considerably different, as latter group is distinguished by the significant presence of European Hg-s; I2a1a2b-L621, R1b1a1b1a1a1-U106 and the Finno-Permic N1a1a1a1a2-Z1936 branch. Their Siberian N1a1a1a1a4 subclade also points at different source populations among ancestors of Yakuts, Evenks and Evens. Nevertheless the east Eurasian R1a subclade, R1a1a1b2a-Z94 seems to be a common element of the Hun, Avar and Conqueror elite. In contrast to Avars, all three Hun lineages have parallels among the Conquerors, but strong inferences cannot be drawn due to small sample size.
It is generally accepted that the Hungarian language was brought to the Carpathian Basin by the Conquerors. Uralic speaking populations are characterized by a high frequency of Y-Hg N, which have often been interpreted as a genetic signal of shared ancestry. Indeed, recently a distinct shared ancestry component of likely Siberian origin was identified at the genomic level in these populations, modern Hungarians being a puzzling exception35. The Conqueror elite had a significant proportion of N Hgs, 7% of them carrying N1a1a1a1a4-M2118 and 10% N1a1a1a1a2-Z1936, both of which are present in Ugric speaking Khantys and Mansis22. At the same time none of the examined Conquerors belonged to the L1034 subclade of Z1936, while all of the Khanty Z1936 lineages reported in36 proved to be L1034 which has not been tested in the22 study. Population genetic data rather position the Conqueror elite among Turkic groups, Bashkirs and Volga Tatars, in agreement with contemporary historical accounts which denominated the Conquerors as “Turks”37. This does not exclude the possibility that the Hungarian language could also have been present in the obviously very heterogeneous, probably multiethnic Conqueror tribal alliance.
Scientific Reports volume 9, Article number: 16569 (2019)
|
|
|
|
Post by Admin on Feb 15, 2020 21:35:12 GMT
Abstract For millennia, the Pontic-Caspian steppe was a connector between the Eurasian steppe and Europe. In this scene, multidirectional and sequential movements of different populations may have occurred, including those of the Eurasian steppe nomads. We sequenced 35 genomes (low to medium coverage) of Bronze Age individuals (Srubnaya-Alakulskaya) and Iron Age nomads (Cimmerians, Scythians, and Sarmatians) that represent four distinct cultural entities corresponding to the chronological sequence of cultural complexes in the region. Our results suggest that, despite genetic links among these peoples, no group can be considered a direct ancestor of the subsequent group. The nomadic populations were heterogeneous and carried genetic affinities with populations from several other regions including the Far East and the southern Urals. We found evidence of a stable shared genetic signature, making the eastern Pontic-Caspian steppe a likely source of western nomadic groups. INTRODUCTION The Pontic-Caspian steppe (PCS), stretching from the southern Urals to the western North Pontic lands, was the stage of various demographic changes in the past, and several of those remain unknown. During the Bronze and Iron Age, the area was inhabited by a succession of nomadic populations that had significant impact on the cultural development of both Asia and Europe (1, 2). Possibly the best known of these groups is the Yamnaya. Recent genomic studies have revealed cross-continental Early Bronze Age migrations (~3000 BCE) of the nomadic people associated with the Yamnaya horizon (3, 4). The migration introduced the Caucasus genetic component to the genetic landscape of Europe. In Central Europe, Yamnaya ancestry first appeared among people from the Corded Ware complex and has since been found in many subsequent ancient and present-day populations. However, the Pontic-Caspian steppe was critical not only for Early Bronze Age Yamnaya migrations but also because of succeeding movements and population transformations that took place in the developed classical stage of the Late Bronze and Iron Ages between 1800 BCE and 400 CE. This period covered the development of the Srubnaya and Alakulskaya Cultures (~1800–1200 BCE), associated with small settlement sites distributed from the Urals to the Dnieper valley (1). From around 1000 BCE, pre-Scythian nomadic populations started to appear in the western Pontic-Caspian steppe including the Cimmerians known from historical sources (5). Despite regional variation and local peculiarities, the Cimmerians were not associated with any uniform type of archaeological material culture (6). In the seventh century BCE, they were succeeded by the Scythians, who plausibly pushed the Cimmerians into Asia Minor (7). Between 700 and 300 BCE, the Scythians, representing mobile pastoral nomads of a new militaristic type (1), dominated the Pontic-Kazakh steppe, occupying an area from the Altai to the Carpathian Mountains. Their decline began around 300 BCE and was caused by intensifying hostile relations with the Macedonians in the West and the invasion of the Sarmatians from the East. The Sarmatians and the Scythians are thought to have coexisted for a few centuries, but eventually, the former group prevailed (2), resulting in the Scythian downfall. The Sarmatians are believed to comprise a number of groups of similar nomadic background (8), and they became the politically most influential force within the eastern fringes of the Roman Empire at the time. Their decline (~400 CE) was associated with the attack of the Goths and the subsequent invasion of the Huns (8). The genomic structure of the Bronze and Iron Age (1800 BCE–400 CE) populations in the Pontic-Caspian steppe has not been fully resolved. While earlier genomic studies have suggested close links between the Srubnaya and the central European Late Neolithic and Bronze Age populations (9), our knowledge of the genetic origins of the Cimmerians is limited. Genetic analyses of maternal lineages of Scythians suggest a mixed origin and an east-west admixture gradient across the Eurasian steppe (10–12). The genomics of two early Scythian Aldy-Bel individuals (13) showed genetic affinities to eastern populations of Central Asia (12). However, population interactions and the origin of Scythians of the Pontic-Caspian steppe remain poorly understood. Similarly, little is known about the origins and genetic affinities of the Sarmatians. Genomic studies suggest that the latter group may have been genetically similar to the eastern Yamnaya and Poltavka Bronze Age groups (12). To investigate the demographic dynamics in the Pontic-Caspian steppe, we generated and analyzed genomes of the Late Bronze and Iron Age individuals from the region (Fig. 1, A and B).  Fig. 1 Radiocarbon ages and geographical locations of the ancient samples used in this study. Figure panels presented counterclockwise: (A) Bar plot visualizing approximate timeline of presented and previously published individuals. (B) Map showing the locations of ancient individuals sequenced in this study and the locations of previously published ancient individuals used in comparative analyses. (C) Principal component analysis (PCA) plot visualizing 35 Bronze Age and Iron Age individuals presented in this study and in published ancient individuals (table S5) in relation to modern reference panel from the Human Origins data set (41). RESULTS We produced genome-wide sequence data with genome coverage between 0.01× and 2.9× per individual for 35 Bronze Age and Iron Age individuals from the Pontic-Caspian steppe from four chronologically sequential cultural groups, which comprise Srubnaya-Alakulskaya individuals (n = 13), Cimmerians (n = 3), Scythians (n = 14), and Sarmatians (n = 5), with radiocarbon dates between ca. 1900 BCE and 400 CE (Fig. 1, A and B; tables S1 to S3; and fig. S1, A and B). All DNA libraries displayed damage patterns typical of ancient DNA (fig. S2) (14). To ensure data integrity, we calculated mitochondrial DNA (mtDNA)–based contamination levels using distribution of private polymorphisms in mtDNA (15) and a Bayesian likelihood method (16). The former yielded point estimates of contamination between 0 and 10% [95% confidence intervals (CIs) between 0 and 17%], and the latter method revealed that all individuals carried sequences with >89% probability of being authentic (table S4). Thus, we included all sequenced individuals in the downstream analyses. Late Bronze Age (LBA) Srubnaya-Alakulskaya individuals carried mtDNA haplogroups associated with Europeans or West Eurasians (17) including H, J1, K1, T2, U2, U4, and U5 (table S3). In contrast, the Iron Age nomads (Cimmerians, Scythians, and Sarmatians) additionally carried mtDNA haplogroups associated with Central Asia and the Far East (A, C, D, and M) (table S3) (11, 18). The absence of East Asian mitochondrial lineages in the more eastern and older Srubnaya-Alakulskaya population suggests that the appearance of East Asian haplogroups in the steppe populations might be associated with the Iron Age nomads, starting with the Cimmerians. The Y chromosome haplogroup variation in 17 of 18 males was limited to two major haplogroup lineages within the macrohaplogroup “R” (table S3). The Srubnaya-Alakulskaya individuals carried the Y haplogroup R1a, which showed a major expansion during the Bronze Age (19). It has previously been found in Bronze Age individuals from the Krasnoyarsk Kurgan in Siberia (20). The Iron Age nomads mostly carried the R1b Y haplogroup, which is characteristic of the Yamnaya of the Russian steppe (4). An exception was a Cimmerian individual (cim358) who carried the Q1* lineage associated with the east (table S3). Genetic relationships between Eurasian steppe nomads and present-day populations PCA on the autosomal genomic data (Fig. 1C and table S5) revealed the following: (i) Srubnaya-Alakulskaya individuals exhibited genetic affinity to northern and northeastern present-day Europeans (fig. S3), and these results were also consistent with outgroup f3 statistics (table S6 and fig. S4A). (ii) The Cimmerian individuals, representing the time period of transition from Bronze to Iron Age, were not homogeneous regarding their genetic similarities to present-day populations according to the PCA. F3 statistics confirmed the heterogeneity of these individuals in comparison with present-day populations (table S6 and figs. S3 and S4C). (iii) The Scythians reported in this study, from the core Scythian territory in the North Pontic steppe (12), showed high intragroup diversity. In the PCA, they are positioned as four visually distinct groups compared to the gradient of present-day populations (Fig. 1C): (i) A group of three individuals (scy009, scy010, and scy303) showed genetic affinity to north European populations, hereafter referred to as a north European (NE) cluster. (ii) A group of four individuals (scy192, scy197, scy300, and scy305) showed genetic similarities to southern European populations, hereafter referred to as a south European (SE) cluster. (iii) A group of three individuals (scy006, scy011, and scy193) located between the genetic variation of Mordovians and populations of the North Caucasus, hereafter referred to as a steppe cluster (SC). In addition, one Srubnaya-Alakulskaya individual (kzb004), the most recent Cimmerian (cim357), and all Sarmatians fell within this cluster. In contrast to the Scythians, and despite being from opposite ends of the Pontic-Caspian steppe, the five Sarmatians grouped close together in this cluster. (iv) A group of three Scythians (scy301, scy304, and scy311) formed a discrete group between the SC and SE and had genetic affinities to present-day Bulgarian, Greek, Croatian, and Turkish populations, hereafter referred to as a central cluster (CC). All PCA results were consistent with outgroup f3 statistics (table S6 and figs. S3 and S4, B and D). Finally, one individual from a Scythian cultural context (scy332) is positioned outside of the modern West Eurasian genetic variation (Fig. 1C) but shared genetic drift with East Asian populations (table S6 and fig. S4B). |
|
|
|
Post by Admin on Feb 16, 2020 6:40:46 GMT
Genetic relationships between Srubnaya-Alakulskaya and other ancient populations The Bronze Age in Eurasia was a dynamic period with several human groups participating in different migratory processes. As a result, there were extensive interactions between European Bronze Age populations (3). The Srubnaya-Alakulskaya individuals, originating from a site of cultural dualism in the forest steppe of the Trans-Volga region, were genetically similar to the previously published Srubnaya and Andronovo individuals from the Pontic-Kazakh steppe (3, 9) and to the European Bronze Age groups, including individuals of the Corded Ware, Unetice, and Bell Beaker complexes (Fig. 1C and fig. S5). Consistent with other Bronze Age populations, the Srubnaya-Alakulskaya individuals were positioned between the genetic variation of the European Mesolithic and the Near East Neolithic populations, being closer to the former and especially to the east European hunter-gatherers (Fig. 1C and figs. S6 and S8). These individuals had higher genetic affinity to Scythians compared to other Iron Age groups (fig. S9). To test whether our Bronze Age sample from a cultural mixing zone between the Srubnaya and the Alakulskaya complexes shared more genetic drift with previously published representatives of Srubnaya, we calculated f4 statistics of f4(Yoruba, SrubnayaX, SrubnayaY, BronzeX), where “BronzeX” refers to a Bronze Age Russian population. The test revealed that the Srubnaya-Alakulskaya population formed a clade together with Andronovo, Afanasievo, and Sintashta to the exclusion of the previously published data of other Srubnaya individuals (tables S7 and S8). Furthermore, to trace the plausible origin the Caucasus genetic component identified in Srubnaya-Alakulskaya individuals, we adopt the f4 statistics in the form of f4(Yoruba, Srubnaya-Alakulskaya, PopulationX, Yamnaya), where PopulationX was one of the Eurasian Bronze Age populations. The results showed that Srubnaya-Alakulskaya formed a clade together with Yamnaya to the exclusion of other Bronze Age populations from Russia, Armenia, Jordan, and Hungary. This finding indicates that the Caucasus genetic contribution to the Srubnaya-Alakulskaya individuals was mediated by steppe ancestry instead of originating from the Levant (table S9). Both mean f3 statistics within populations and conditional nucleotide diversity (21) revealed that the genetic diversity was highest in the LBA Srubnaya-Alakulskaya population from the southern Ural region compared to all other Eurasian Bronze Age populations (Fig. 2, A and B, and table S10).  Fig. 2 Genetic diversity and ancestral components of Srubnaya-Alakulskaya population. Diversity and ancestral components of Srubnaya-Alakulskaya population (here called “Srubnaya”): (A) Mean f3 statistics for Srubnaya and other Bronze Age populations. Srubnaya group was color-coded the same as with PCA. (B) Pairwise mismatch estimates for Bronze Age populations. (C) ADMIXTURE results for K = 15. K = 15 was chosen to display since it shows SA component (lilac) and Northeast Asian (NEA, “dark green”) components in addition to the other components. BA, Bronze Age; EBA, Early Bronze Age; MBA, Middle Bronze Age; CA, Chalcolithic; N, Neolithic; EN, Early Neolithic; LN, Late Neolithic; HG, hunter-gatherers; SHG, Scandinavian hunter-gatherers (fig. S10). To evaluate the ancestral components of Srubnaya-Alakulskaya, we conducted ADMIXTURE analysis for K = 2 to K = 15 ancestral clusters (fig. S10A). K = 15 clusters revealed that Srubnaya-Alakulskaya individuals consisted of two major ancestral components; first, an “orange” component predominantly found in west and NE hunter-gatherers (WHG) and in present-day NE populations, and second, a “light green” component typical of Caucasus hunter-gatherers (CHG) found in south Asian (SA) modern populations. The component associated with Neolithic groups and present-day Near Eastern populations (NEN, “dark red”) contributed less to our Srubnaya-Alakulskaya individuals compared to the European Bronze Age populations (Fig. 2C and fig. S10B). Genetic relationships between the Iron Age nomads and other ancient populations In the 10th century BCE, during the transition from the Bronze Age to the Iron Age, the Cimmerians appear in the Pontic-Caspian steppe. In the PCA, the chronologically youngest Cimmerian individual (cim357) grouped within the SC including all Sarmatians, one Srubnaya-Alakulskaya individual, and three Scythian individuals. The other Cimmerian individuals were positioned in close proximity to a number of eastern Iron Age individuals from the Altai region (3), as well as Aldy-Bel and Zevakino-Chillikta individuals of the Altai and Western Mongolia regions (Fig. 1C and fig. S9) (12). We tested whether Cimmerians formed a clade with the subsequent Iron Age populations including Scythians and Sarmatians, to the exclusion of Bronze Age populations by calculating f4 statistics in the form of f4(Yoruba, Cimmerians; Srubnaya&Scythians&Sarmatians, BronzeAgePopulation), showing that the Cimmerians shared more drift with the Bronze Age populations of Russia compared to the Srubnaya/-Alakulskaya but not compared to Scythians or Sarmatians (Fig. 3A and tables S11, S12, and S16). The Cimmerians shared more drift with the far eastern Karasuk population compared to the geographically more closely located Srubnaya/-Alakulskaya population (table S16), corroborating the existence of the “Karasuk-Cimmerian cultural-historical community” (22). Pairwise mismatch estimates revealed a slightly higher genetic diversity in Cimmerians compared to that of the Srubnaya-Alakulskaya population (table S10). ADMIXTURE analysis (K = 15) revealed that Cimmerian individuals carried predominantly the WHG and CHG components similar to the Srubnaya individuals, but they also carried other ancestral components including NEN (dark red), Northeast Asian (NEA) (“dark green”), and Southeast Asian (SEA, “light blue”) components. From the oldest to the most recent sample, the amount of NEN component increased, while the NEA and SEA components decreased through time in the Cimmerians (Fig. 3D and fig. S10). The admixture results were further tested with f3 statistics, which were consistent with the observed pattern by means of shared genetic drift between the Cimmerians and the tested populations used as proxies for different genetic components identified in admixture analyses (table S13). These results implicate a more eastern origin of the Cimmerians compared to that of the Srubnaya-Alakulskaya population and an increased amount of NEN-originated admixture in these individuals through time. |
|







