|
|
Post by Admin on Jul 5, 2019 18:02:59 GMT
DNA sequencing of ten Philistine skeletons suggests they really were a genetically distinct community. Around 1200 BC, in at least one key Philistine city there was an influx of south European genes, suggesting a surge of Greek immigrants to the region, says Michal Feldman at the Max Planck Institute for the Science of Human History in Jena, Germany.  The Bible’s Old Testament makes numerous references to the Philistines; for instance, Goliath, the “giant” who fought David, was a Philistine, as probably was Delilah, said to have betrayed Samson by cutting his hair. Multiple excavations from sites of ancient Philistine cities, such as Ashkelon, on the coast of what is now Israel, have yielded pottery remains that are Greek in style. One suggestion was that people could simply have adopted Aegean cultural practices via sea trading routes.  To investigate, Feldman’s team tried to extract DNA from 108 skeletal remains excavated from various burial places in Ashkelon that had been dated to either the Bronze Age or Iron Age. Ten produced useful genetic information from their bones or teeth, and this was compared with DNA from other populations all over the world, both ancient and modern. The remains could be divided into three time periods. The earliest three individuals found in a necropolis came from about 1600 BC, four were infants that had been buried under houses around 1200 BC, and three more individuals were from a cemetery by the city wall and came from about 1100 BC.  The people from the middle period had significant ancestry from southern Europe, with 20 to 60 per cent similarity to DNA from ancient skeletons from Crete and Iberia and that from modern people living in Sardinia, an Italian island. However, the last group of three bodies had no more detectable Greek ancestry than the first group. “Probably all these immigrants that came in intermarried with the local population until this foreign ancestry was diluted,” says Feldman.  “Putting the genetic data together with the archaeological data strengthens the case that there was migration from the areas that we now call Greece and western Turkey,” says Christoph Bachhuber of the University of Oxford, who wasn’t involved in the work. |
|
|
|
Post by Admin on Jul 6, 2019 17:39:36 GMT
Within the history of the Eastern Mediterranean, the end of the Bronze Age and the beginning of the Iron Age were marked by exceptional cultural disarray that followed the demise of prosperous economies and cultures in Greece, Egypt, the Levant, and Anatolia (1). During the 12th century BCE, coincident with these events, substantive cultural changes appeared in the archeological record of Ashkelon, Ashdod, and Ekron, three of the five core cities mentioned as “Philistine” in the Hebrew bible (2–4). These settlements were distinct from neighboring sites in architectural traditions and material culture (2–4). Resemblances between the new cultural traits and 13th century patterns found in the Aegean have led some scholars to explain this so-called “Philistine phenomenon” by a migration from an Aegean-related source, potentially associated with the “Sea Peoples,” a population that is thought to have settled in other parts of the coastal Eastern Mediterranean (2, 3). This hypothesis has been challenged by those arguing that this cultural change was driven by a diffusion of knowledge or internal development of ideas (5–8) rather than by a large-scale movement of people. Even for those who do accept the idea of large-scale mobility, the homeland of the new arrivals is disputed with suggested alternatives including Cyprus or Cilicia (4), a mixture of non-Aegean east Mediterranean peoples (8), and mixed heterogeneous maritime groups, akin to pirates (9). Proposed links go as far as northern Italy where depopulation events have been suggested to trigger population movements throughout the Mediterranean (10). Recent ancient DNA (aDNA) studies have reported a high degree of genetic continuity in the Levant during the late Pleistocene and early Holocene that was followed by increasing population admixtures with Anatolian- and Iranian-related populations at least up to the Middle Bronze Age (11–14). Genome-wide data from Late Bronze and Iron Age populations have, so far, not been available for this region.  Fig. 1 Overview of locations and ages of analyzed individuals. (A) Locations of newly reported and other selected published genomes. The newly reported Ashkelon populations are annotated in the upper corner. (B) The range of average ages of the ancient groups is given in thousands of years (ka) BCE. Here, we report genome-wide data from human remains excavated at the ancient seaport of Ashkelon, forming a genetic time series encompassing the Bronze to Iron Age transition (Fig. 1, A and B). We find that all three Ashkelon populations derive most of their ancestry from the local Levantine gene pool. The early Iron Age population was distinct in its high genetic affinity to European-derived populations and in the high variation of that affinity, suggesting that a gene flow from a European-related gene pool entered Ashkelon either at the end of the Bronze Age or at the beginning of the Iron Age. Of the available contemporaneous populations, we model the southern European gene pool as the best proxy for this incoming gene flow. Last, we observe that the excess European affinity of the early Iron Age individuals does not persist in the later Iron Age population, suggesting that it had a limited genetic impact on the long-term population structure of the people in Ashkelon. Genome-wide data across the Bronze/Iron Age transition in Ashkelon We extracted and sequenced DNA from 108 skeletal elements excavated in Ashkelon. In line with the low DNA preservation previously reported for the southern Levant (11–14), only 10 yielded sufficient amounts of human DNA (data file S1). Sequencing libraries for these 10 individuals were enriched for human DNA using an established in-solution DNA capture targeting 1.24 million genome-wide single-nucleotide polymorphisms (SNPs) (“1240 k capture”) (15–17). The earliest three individuals were excavated from a Bronze Age necropolis (18–20) and dated by the archeological context to the Middle Bronze IIC–Late Bronze Age II (labeled ASH_LBA; two of the individuals were directly carbon-dated to 1746 to 1542 cal BCE). Four early Iron Age infants were recovered from burials beneath late 12th century Iron Age I houses (labeled ASH_IA1; directly carbon-dated to 1379 to 1126 cal BCE), and three later Iron Age individuals were recovered from a cemetery adjacent to the city wall of ancient Ashkelon, which was estimated to have been used between the 10th and the 9th century BCE (labeled ASH_IA2; one individual directly carbon-dated to 1257 to 1042 cal BCE) (Fig. 1, A and B, Table 1, table S1, and text S1). While the chronological ranges estimated by the archeological context are approximately within the range of the radiocarbon assays conducted directly on the petrous bones sampled for DNA (table S1), we caution that they should be taken as a rough estimate, since the presence of marine carbon in the diet could make the calibrated radiocarbon dates older than the true age.  Fig. 2 PCA and ADMIXTURE analysis. (A) Ancient genomes (marked with color-filled symbols) projected onto the principal components inferred from present-day west Eurasians (gray circles) (fig. S2). The newly reported Ashkelon populations are annotated in the upper corner. (B) ADMIXTURE analysis. A selected set of ancient individuals (as well as present-day Sardinians) is plotted (K = 9 was chosen since it is the cluster number that maximizes components correlated to the most differentiated populations in the west Eurasian PCA). To formally test the qualitative observations based on the PCA and ADMIXTURE analyses, we used f4-statistics (24). Consistent with the ASH_LBA positions on PC2, the f4-statistic of the form f4 (ASH_LBA, Levant_N/Levant_ChL; test, Mbuti) (figs. S4 and S5) estimated excess allele sharing between ASH_LBA and ancient Iranian/Caucasus-related populations (such as CHG, Iran_N, and Iran_ChL) compared to the Neolithic/Chalcolithic Levantines (Levant_N and Levant_ChL), confirming previous reports of post-Neolithic gene flows from prehistoric populations related to Iran or the Caucasus into the Levant (11–13). Accordingly, modeling ASH_LBA as a two-way admixture using qpAdm (12, 17) produced a fitting model (χ2P = 0.445), in which ASH_LBA derives around 60% of their ancestry from Levant_ChL (60.0 ± 6.5%; estimate ±1 SE) and the rest from Iran_ChL. The spatiotemporal origins of this post-Neolithic gene flow remain broadly defined since alternative two-way models also fit, albeit with smaller P values. In these models, Anatolia_EBA/Anatolia_BA/Armenia_ChL are used as the second source, replacing Iran_ChL (χ2P = 0.053 to 0.060; table S3). We then examined whether there are noticeable genetic differences between ASH_LBA and the earlier Bronze Age Levant populations by measuring f4 (ASH_LBA, Jordan_EBA/Lebanon_MBA; test, Mbuti (figs. S6 and S7). We find that ASH_LBA and both earlier Bronze Age populations are symmetrically related to all test populations within our data’s resolution (|f4| < 3 SE). However, we observe a marginal excess of affinity between ASH_LBA and populations genetically related to ancient Iran and the Caucasus (such as CHG and Steppe_Eneolithic) when compared to Jordan_EBA (fig. S7). Within our current resolution, we cannot distinguish whether this affinity is due to a small-scale gene flow from an Iranian/Caucasus-related population entering the Levant between the Early and Late Bronze Age or whether it reflects a certain population structure during this period. Nonetheless, the apparent symmetry suggests a high degree of genetic continuity throughout at least a millennium between culturally and geographically distinct Bronze Age Levantine groups occupying a region that stretches from the inland southern Levant where present-day Jordan is located and along the coastal regions of present-day Israel and Lebanon. |
|
|
|
Post by Admin on Jul 7, 2019 17:42:05 GMT
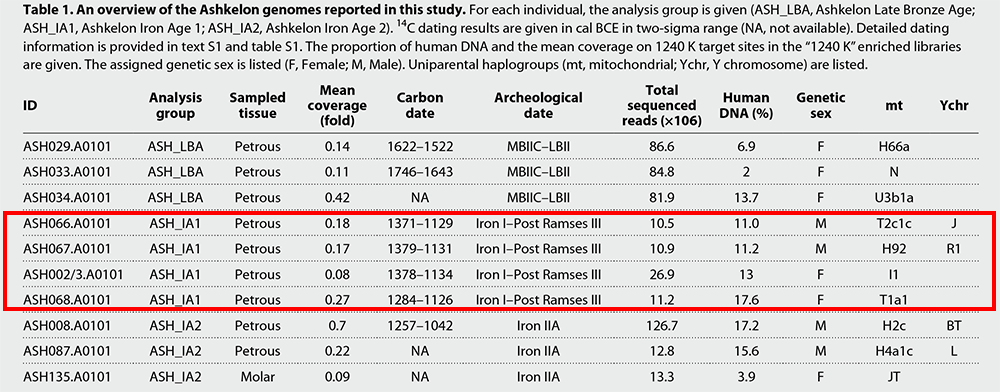 Table 1 An overview of the Ashkelon genomes reported in this study. For each individual, the analysis group is given (ASH_LBA, Ashkelon Late Bronze Age; ASH_IA1, Ashkelon Iron Age 1; ASH_IA2, Ashkelon Iron Age 2). 14C dating results are given in cal BCE in two-sigma range (NA, not available). Detailed dating information is provided in text S1 and table S1. The proportion of human DNA and the mean coverage on 1240 K target sites in the “1240 K” enriched libraries are given. The assigned genetic sex is listed (F, Female; M, Male). Uniparental haplogroups (mt, mitochondrial; Ychr, Y chromosome) are listed. Genetic discontinuity between the Bronze Age and the early Iron Age people of Ashkelon In comparison to ASH_LBA, the four ASH_IA1 individuals from the following Iron Age I period are, on average, shifted along PC1 toward the European cline and are more spread out along PC1, overlapping with ASH_LBA on one extreme and with the Greek Late Bronze Age “S_Greece_LBA” (25) on the other (Fig. 2A). Similarly, genetic clustering assigns ASH_IA1 with an average of 14% contribution from a cluster maximized in the Mesolithic European hunter-gatherers labeled “WHG” (shown in blue in Fig. 2B) (15, 22, 26). This component is inferred only in small proportions in earlier Bronze Age Levantine populations (2 to 9%). In light of these observations, we formally tested the variation within each of the three Ashkelon populations by computing the variance of the square rooted statistic f4 (Ashkelon individual 1, Ashkelon individual 2; test, Mbuti) (fig. S8). Consistent with the wide spread of ASH_IA1 in the west Eurasian PCA, the variance is significantly higher in ASH_IA1 than in either ASH_LBA or ASH_IA2 (P < 2.2 × 10−16 for both by a Fligner-Killeen test) (table S4 and fig. S8), suggesting that the ASH_IA1 individuals were more heterogeneous in their genetic affinities to global populations in respect to both ASH_LBA and ASH_IA2. We next formally quantified the genetic differences between ASH_IA1 and the earlier ASH_LBA by calculating f4 (ASH_IA1, ASH_LBA; test, Mbuti) (Fig. 3A). In agreement with the PCA and ADMIXTURE results, only European hunter-gatherers (including WHG) and populations sharing a history of genetic admixture with European hunter-gatherers (e.g., as European Neolithic and post-Neolithic populations) produced significantly positive f4-statistics (Z ≥ 3), suggesting that, compared to ASH_LBA, ASH_IA1 has additional European-related ancestry. To estimate the levels of the European-related ancestry in all Ashkelon populations, we compared qpAdm models of two-way mixtures (Levant_ChL and Iran_ChL; i.e., 0% contribution from WHG) to three-way ones, in which we add WHG as the third source (Fig. 3C and table S5). ASH_LBA and ASH_IA2 fit well with the two-way model (χ2P = 0.445 and χ2P = 0.313, respectively; table S5), whereas the three-way one infers small nonsignificant proportions (−2.3 ± 2.2 and 2.0 ± 2.2% for ASH_LBA and ASH_IA2, respectively; table S5). In contrast, for ASH_IA1, the two-way model is inadequate (χ2P = 3.80 × 106; table S5) and the three-way one fits (χ2P = 0.765). Thus, of the three, additional WHG-like ancestry is only necessary to model ASH_IA1.  Fig. 3 European-related admixture detected in ASH_IA1. (A) ASH_IA1 shares access affinity with European-related populations compared to ASH_LBA. We plot the top and bottom 40 values of f4 (ASH_IA1, ASH_IA2; X, Mbuti) on the map. Circles mark the ancient populations and triangles the present-day ones. Z-scores calculated by 5-centimorgan block jackknifing are represented by the size of the symbols. “X” share more alleles with ASH_IA1 when values are positive and with ASH_IA2 when negative. The five groups with the most positive values are annotated on the map (Z > 2.3). (B) We plot the ancestral proportions of the Ashkelon individuals inferred by qpAdm using Iran_ChL, Levant_ChL, and WHG as sources ±1 SEs. P values are annotated under each model. In cases when the three-way model failed (χ2P < 0.05), we plot the fitting two-way model. The WHG ancestry is necessary only in ASH_IA1. We find that the PC1 coordinates positively correlate with the proportion of WHG ancestry modeled in the Ashkelon individuals (fig. S9 and table S6), suggesting that WHG reasonably tag a European-related ancestral component within the ASH_IA1 individuals. However, these Mesolithic individuals are unlikely to be a good proxy for the true source in the much later early Iron Age. To examine more proximate sources, we compiled a set of chronologically and geographically relevant candidate populations, including populations that shared higher affinity with ASH_IA1 compared to ASH_LBA in the above f4-statistic. Subsequently, we modeled ASH_IA1 as two-way and three-way mixtures of ASH_LBA and combinations of the candidate populations (table S7). Of the 51 tested models, we find four plausible ones (χ2P > 0.05), all are two-way mixtures. The best supported one (χ2P = 0.675) infers that ASH_IA1 derives around 43% of ancestry from the Greek Bronze Age “Crete_Odigitria_BA” (43.1 ± 19.2%) and the rest from the ASH_LBA population. ASH_IA1 could also be modeled with either the modern “Sardinian” (35.2 ± 17.4%; χ2P = 0.070), the Bronze Age “Iberia_BA” (21.8 ± 21.1%; χ2P = 0.205), or the Bronze Age “Steppe_MLBA” (15.7 ± 9.1%; χ2P = 0.050) as the second source population to ASH_LBA. To check whether these results are due to the low coverage of ASH_LBA, we repeated this analysis, but this time, we modeled ASH_IA1 as a three-way mixture of each of the candidate populations, Levant_ChL and Iran_ChL. The two latter populations have higher genome coverage and can model ASH_LBA well in combination (table S3). In this analysis, only the models including “Sardinian,” “Crete_Odigitria_BA,” or “Iberia_BA” as the candidate population provided a good fit (χ2P = 0.715, 49.3 ± 8.5%; χ2P = 0.972, 38.0 ± 22.0%; and χ2P = 0.964, 25.8 ± 9.3%, respectively). We note that, because of geographical and temporal sampling gaps, populations that potentially contributed the “European-related” admixture in ASH_IA1 could be missing from the dataset. Therefore, better proxies might be found in the future when more data is available. Nonetheless, the tested candidate populations from Anatolia, Egypt, and the Levant that did not produce well-fitting models can be excluded as potential sources of the admixture observed in ASH_IA1. The transient impact of the “European-related” gene flow on the Ashkelon gene pool The ASH_IA2 individuals are intermediate along PC1 between the ASH_LBA ones and the earlier Bronze Age Levantines (Jordan_EBA/Lebanon_MBA) in the west Eurasian PCA (Fig. 2A). Notably, despite being chronologically closer to ASH_IA1, the ASH_IA2 individuals position closer, on average, to the earlier Bronze Age individuals. Such a reduced affinity to the European populations is also apparent in the genetic clustering results, showing that the European hunter-gatherer–related ancestry contributes considerably less to ASH_IA2 than to ASH_IA1 (8 and 14%, respectively; Fig. 2B). To test for differences in affinities between ASH_IA2 and ASH_IA1, we measured the f4-statistic of the form f4 (ASH_IA2, ASH_IA1; test, Mbuti). This statistic produced no significantly positive results, and all significantly negative statistics (Z ≤ −3) are with test populations from either Europe or Anatolia (fig. S10). This, again, highlights the increased European-related affinity of ASH_IA1.  The transient excess of European-related genetic affinity in ASH_IA1 can be explained by two scenarios. The early Iron Age European-related genetic component could have been diluted by either the local Ashkelon population to the undetectable level at the time of the later Iron Age individuals or by a gene flow from a population outside of Ashkelon introduced during the final stages of the early Iron Age or the beginning of the later Iron Age. Considering the symmetry measured between ASH_IA2 and ASH_LBA in the f4-statistic of the form f4 (ASH_IA2, ASH_LBA; test, Mbuti) (|Z| < 2.8; fig. S11), the replacing population is likely to have stemmed from the Late Bronze Age Levantine gene pool, whether or not from Ashkelon. By modeling ASH_IA2 as a mixture of ASH_IA1 and earlier Bronze Age Levantines/Late Period Egyptian, we infer a range of 7 to 38% of contribution from ASH_IA1, although no contribution cannot be rejected because of the limited resolution to differentiate between Bronze Age and early Iron Age ancestries in this model (table S8). DISCUSSION By investigating genome-wide data from Ashkelon, we address long-pending historical questions regarding the demographic developments underlying the Late Bronze Age to Iron Age cultural transformation. On a larger regional scale, these data begin to fill a temporal gap in the genetic map of the southern Levant, revealing persistence of the local Levantine gene pool throughout the Bronze Age for over a millennium. At the same time, by the “zoomed-in” comparative analysis of the Ashkelon genetic time transect, we find that the unique cultural features in the early Iron Age are mirrored by the distinct genetic composition we detect in ASH_IA1. Our analysis suggests that this genetic distinction is due to a European-related gene flow introduced in Ashkelon during either the end of the Bronze Age or the beginning of the Iron Age. This timing is in accord with estimates of the Philistines arrival to the coast of the Levant, based on archeological and textual records (2–4). We find that, within no more than two centuries, this genetic footprint introduced during the early Iron Age is no longer detectable and seems to be diluted by a local Levantine-related gene pool. The relatively rapid disappearance of this signal stresses the value of temporally dense genetic sampling for addressing historical questions. Transient gene flows, such as the one detected here, might be overlooked because of a lack of representative samples, potentially leading to erroneous conclusions. In geographic regions unfavorable to DNA preservation, obtaining such datasets requires exhaustive sampling and the utilization and further development of advanced technologies such as DNA enrichment techniques (15–17) and targeted sampling strategies (27). We do not rule out that some gene flow occurred during the Bronze Age as low significance of the f4-statistics might be due to the limited statistical power of our data stemming from either insufficient coverage or a lack of appropriate contemporaneous proxy populations. Thus, additional sampling is needed to further investigate the question of the genetic diversity within the Levantine Bronze Age populations and to characterize the spatiotemporal extent of potential incoming gene flows. Similarly, a larger sample size might help to accurately infer the extent and magnitude of the early Iron Age gene flows and to identify more precisely the populations introducing the European-related component to Ashkelon. While our modeling suggests a southern European gene pool as a plausible source, future sampling in regions such as Cyprus, Sardinia, and the Aegean, as well as in the southern Levant, could better resolve this question. Science Advances 03 Jul 2019: Vol. 5, no. 7, eaax0061 |
|
|
|
Post by Admin on Jul 16, 2019 22:30:44 GMT
Last weekend, Prime Minister of Israel Benjamin Netanyahu, or whoever in his administration operates his Twitter account, tweeted about a new study that had been published in the journal Science Advances and covered widely in the media, including in Smithsonian. The study analyzed DNA from ten individuals who had been buried at Ashkelon, a coastal city in Israel, between the Bronze Age and Iron Age. The results suggested that the appearance of new genetic signatures in four of the individuals coincided with changes in the archaeological record that have been associated with the arrival of the Philistines more than 3,000 years ago. These genetic traits resembled those of ancient people who lived in what is now Greece, Italy and Spain. The authors asserted that these findings supported the idea that the Philistines, a group of people made infamous in the Hebrew Bible as the enemies of the Israelites, originally migrated to the Levant from somewhere in southern Europe, but quickly mixed with local populations. Commenting on the study, Netanyahu wrote: “There’s no connection between the ancient Philistines & the modern Palestinians, whose ancestors came from the Arabian Peninsula to the Land of Israel thousands of years later. The Palestinians’ connection to the Land of Israel is nothing compared to the 4,000 year connection that the Jewish people have with the land.” The logic here for those who had read the study was confusing. The new research had nothing to say about the genetic history of Jews or Palestinians or the connection those modern populations have to the land. (Though the word "Palestinian" comes from "Philistine," Palestinians are not thought of as the descendants of Philistines; it appears that Netanyahu was using this unrelated point to launch into his argument.) “To me it seemed like it just provided another opportunity—even if it's just tangential—to take a swipe at Palestinians,” says Michael Press, an independent scholar who studies the presentation of archaeology in Israel and the occupied Palestinian territories. “It's hard to blame the authors much here since Netanyahu's use of the study really was a non-sequitur.” (The authors of the study did not wish to comment but are preparing a formal response.) Despite evidence that Jews and Palestinians are genetically closely related, Press and others were also torn about even addressing such inaccuracies in Netanyahu’s comments. Tom Booth, a researcher in the ancient genomics laboratory at the Francis Crick Institute in London, worried that picking apart what the prime minister got wrong about the study would suggest that, in an alternate reality, where his interpretation was scientifically sound, Netanyahu would be justified in using such a study to support his claims about Palestinian rights. “You just need to condemn any attempt to use a study on the past in this way,” Booth says. “The way our ancestors were 4,000 years ago does not really bear on ideas of nation or identity, or it shouldn't in modern nation states.” Hakenbeck and other archaeologists believe geneticists have (unwittingly or not) helped fuel these race-obsessed arguments by reviving old ideas about cultural invasions and migrations that many archaeologists abandoned in the 1960s. Early practitioners of archaeology presented the course of human history as “racialized billiard balls crashing into each other,” Wengrow says. They tended to think of different cultures as clearly bounded entities, and if they saw change happening in the types of ceramics or other artifacts being used an archaeological site, they thought it must mean they were looking at evidence of an invasion. Younger generations of archaeologists have tended to favor explanations involving local invention and the spread of ideas. To them, narratives like the Yamnaya invasion feel like a throwback. (Writer Gideon Lewis-Kraus outlined these tensions at length in an article on ancient DNA for the New York Times Magazine earlier this year.)  “What we’re seeing with ancient DNA studies is a return to early 20th-century thinking—that [geneticists] can get a few samples from a few skeletons, call them by a [cultural] name, usually from a historical source, and say these skeletons are these people, and then we talk about their replacement,” says Rachel Pope, a senior archaeologist at the University of Liverpool. “We are fitting what is actually quite an exciting new science into an antiquated understanding of social mechanisms and how they change. It’s very depressing, and it’s very dangerous.” Hakenbeck says the case of the study on DNA from Ashkelon is a good example of how things could go wrong even when the work itself is quite measured and nuanced. The authors of the paper did emphasize in media interviews that ethnicity and genetics were not the same thing, and that their data reflected a complicated world. |
|
|
|
Post by Admin on Jul 25, 2019 18:17:02 GMT
 The Near East, including the Levant, has been central to human prehistory and history from the expansion out of Africa 50–60 thousand years ago (kya),1 through post-glacial expansions2 and the Neolithic transition 10 kya, to the historical period when Ancient Egyptians, Greeks, Phoenicians, Assyrians, Babylonians, Persians, Romans, and many others left their impact on the region.3 Aspects of the genetic history of the Levant have been inferred from present-day DNA,4, 5 but the more comprehensive analyses performed in Europe6, 7, 8, 9, 10, 11 have shown the limitations of relying on present-day information alone and highlighted the power of ancient DNA (aDNA) for addressing questions about population histories.12 Unfortunately, although the few aDNA results from the Levant available so far are sufficient to reveal how much its history differs from that of Europe,13 more work is needed to establish a thorough understanding of Levantine genetic history. Such work is hindered by the hot and sometimes wet environment,12, 13 but improved aDNA technologies including use of the petrous bone as a source of DNA14 and the rich archaeological remains available encouraged us to further explore the potential of aDNA in this region. Here, we present genome sequences from five Bronze Age Lebanese samples and show how they improve our understanding of the Levant’s history over the last five millennia.  During the Bronze Age in the Levant, around 3–4 kya, a distinctive culture emerged as a Semitic-speaking people known as the Canaanites. The Canaanites inhabited an area bounded by Anatolia to the north, Mesopotamia to the east, and Egypt to the south, with access to Cyprus and the Aegean through the Mediterranean. Thus the Canaanites were at the center of emerging Bronze Age civilizations and became politically and culturally influential.15 They were later known to the ancient Greeks as the Phoenicians who, 2.3–3.5 kya, colonized territories throughout the Mediterranean reaching as far as the Iberian Peninsula.16 However, for uncertain reasons but perhaps related to the use of papyrus instead of clay for documentation, few textual records have survived from the Canaanites themselves and most of their history known today has been reconstructed from ancient Egyptian and Greek records, the Hebrew Bible, and archaeological excavations.15 Many uncertainties still surround the origin of the Canaanites. Ancient Greek historians believed their homeland was located in the region of the Persian Gulf,16, 17 but modern researchers tend to reject this hypothesis because of archaeological and historical evidence of population continuity through successive millennia in the Levant. The Canaanite culture is alternatively thought to have developed from local Chalcolithic people who were themselves derived from people who settled in farming villages 9–10 kya during the Neolithic period.15 Uncertainties also surround the fate of the Canaanites: the Bible reports the destruction of the Canaanite cities and the annihilation of its people; if true, the Canaanites could not have directly contributed genetically to present-day populations. However, no archaeological evidence has so far been found to support widespread destruction of Canaanite cities between the Bronze and Iron Ages: cities on the Levant coast such as Sidon and Tyre show continuity of occupation until the present day. aDNA research has the potential to resolve many questions related to the history of the Canaanites, including their place of origin and fate. Here, we sampled the petrous portion of temporal bones belonging to five ancient individuals dated to between 3,750 and 3,650 years ago (ya) from Sidon, which was a major Canaanite city-state during this period (Figures S1 and S2). We extracted DNA and built double-stranded libraries according to published protocols without uracil-DNA glycosylase treatment.18, 19, 20, 21 We sequenced the libraries on an Illumina HiSeq 2500 using 2× 75 bp reads and processed the sequences using the PALEOMIX pipeline.22 We retained reads ≥30 bp and collapsed pairs with minimum overlap of 15 bp, allowing a mismatch rate of 0.06 between the pairs. We mapped the merged sequences to the hs37d5 reference sequence, removed duplicates, removed two bases from the ends of each read, and randomly sampled a single sequence with a minimum quality of ≥20 to represent each SNP. We obtained a genomic coverage of 0.4–2.3× and a mitochondrial DNA (mtDNA) genome coverage of 53–164× (Table 1). Y chromosome genotypes were jointly called across males from the 1000 Genomes Project, present-day Lebanese, and two identified Canaanite males using FreeBayes v.0.9.18.23 A maximum likelihood phylogeny was inferred using RAxML v.8.2.1024 and visualized using iTOL v.3.5.3.25 In order to assess ancient DNA authenticity, we estimated mtDNA and X chromosome contamination26, 27, 28 (Table S1) and restricted some analyses to sequences with aDNA damage patterns29, 30 (Figures S3 and S4), demonstrating that the sequence data we present are endogenous and minimally contaminated. Additionally, we sequenced whole genomes of 99 present-day Lebanese individuals with informed consent to ∼8× coverage on an Illumina HiSeq 2500 using 2× 100 bp reads in a study approved by The Wellcome Trust Sanger Institute’s Human Materials and Data Management Committee (13/010 and 14/072). We merged the low-coverage Lebanese data with four high-coverage (30×) Lebanese samples,31 1000 Genomes Project phase 3 CEU, YRI, and CHB populations,32 and sequence data previously published from regional populations (Egyptians, Ethiopians, and Greeks).1, 31 Raw calls were generated using bcftools (bcftools mpileup -C50 -pm3 -F0.2 -d10000 | bcftools call -mv, version 1.2-239-g8749475) and filtered to include only SNPs with the minimum of two alternate alleles in at least one population and site quality larger than ten; we excluded sites with a minimum per-population HWE and total HWE less than 0.0133 and sites within 3 bp of an indel. The filtered calls were then pre-phased using shapeit (v.2.r790)34 and their genotypes refined using beagle (v.4.1).35 We have previously described the genetic structure in the Lebanese population using array data from ∼1,300 individuals.4 A principal component analysis (PCA) using the 99 sequenced present-day individuals show that they capture the previously described genetic diversity with distinct clusters reflecting the different cultural groups in Lebanon today (Figure S5). |
|
















