|
|
Post by Admin on Mar 30, 2019 18:22:30 GMT
A parietal fragment from Denisova cave BENCE T. VIOLA1, PHILIPP GUNZ2, SIMON NEUBAUER2, VIVIANE SLON3, MAXIM B. KOZLIKIN4, MICHAEL V. SHUNKOV4,5, MATTHIAS MEYER3, SVANTE PÄÄBO3 and ANATOLY P. DEREVIANKO4,6. 1Department of Anthropology, University of Toronto, 2Department of Human Evolution, Max-Planck-Institute for Evolutionary Anthropology, 3Department of Evolutionary Genetics, Max-Planck-Institute for Evolutionary Anthropology, 4Institute of Archaeology and Ethnography, Russian Academy of Sciences, Siberian Branch, 5Department of Archaeology, Novosibirsk State University, 6Department of Archaeology, Altai State University March 28, 2019 The Denisovans are an Asian sister group of Neanderthals, originally described based on ancient DNA from a phalanx fragment from Denisova Cave (Altai Mountains, Russian Federation). Since then, three teeth found in the cave have been identified from this group. Genetic data indicates that this population contributed genes to modern humans across large parts of Asia and in Melanesia, but currently, no fossils from outside Denisova Cave have been clearly attributed to Denisovans. One reason for this is that the extremely limited morphological evidence hampers comparisons. A newly discovered parietal fragment, Denisova 13, attributed to the Denisovans based on its mtDNA, gives us a first glimpse at the Denisovan cranial morphology. Denisova 13 derives from the South Gallery of Denisova cave, although due to a collapse of the section, its stratigraphic position is not secure. Based on its preservation and the accompanying sediments we tentatively attribute it to Layer 22.The specimen consists of two adjoining fragments of the posterior half of the left parietal, extending about 78 mm laterally of lambda, and about 51 mm anteriorly, preserving portions of both the sagittal and lambdoid sutures. 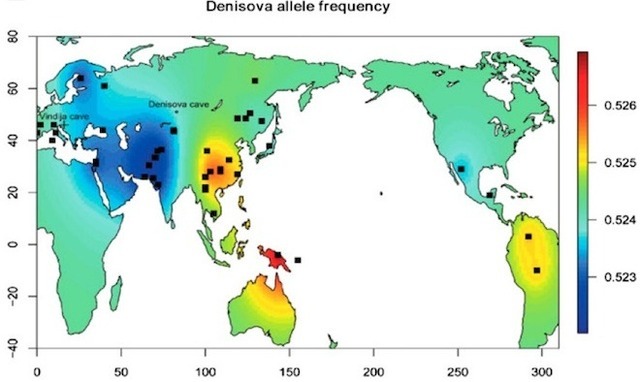 We compare Denisova 13 to a large sample of recent and fossil modern humans, Neanderthals and Middle Pleistocene Homo, using geometric morphometrics based on three-dimensional landmarks and sliding semilandmarks collected on computed-tomographic scans and surface scans. We will discuss the implications of these comparisons to our understanding of Middle and Late Pleistocene hominins in Asia. Funding: Social Sciences and Humanities Research Council (Insight 430-2016-00590 to BV), European Research Council (694707 to SP), the Max-Planck-Society, and the Russian Science Foundation (14-50-00036 to MVS, APD and MBK).  Discovered in two pieces in Siberia’s Denisova Cave in August 2016, the find joins only a handful of fragmentary fossils from these mysterious, extinct hominids. Mitochondrial DNA, a type of genetic material typically inherited from the mother, extracted from the skull pegged it as Denisovan, paleoanthropologist Bence Viola said March 28 at the annual meeting of the American Association of Physical Anthropologists. Viola’s presentation was one of several at the meeting that raised new questions about these Neandertal relatives — including how recently they existed — who are known only from previous discoveries in Denisova Cave. A decade ago, a tiny part of a finger bone yielded Denisovan DNA that proved crucial to identifying the Stone Age population. Sediment analyses indicate that Denisovans periodically inhabited Denisova Cave from around 300,000 to 50,000 years ago, with Neandertals reaching the cave after around 200,000 years ago (SN: 3/2/19, p. 11). But little else is known about Denisovans’ evolutionary history or identity. It’s long been unclear, for instance, if Denisovans belonged to a distinct Homo species. And most researchers say that the new evidence is still not enough to resolve that mystery. “We’re a long way from solving the species question about Denisovans,” said paleoanthropologist Chris Stringer of the Natural History Museum in London, who gave a talk at the meeting on what’s known about the group. 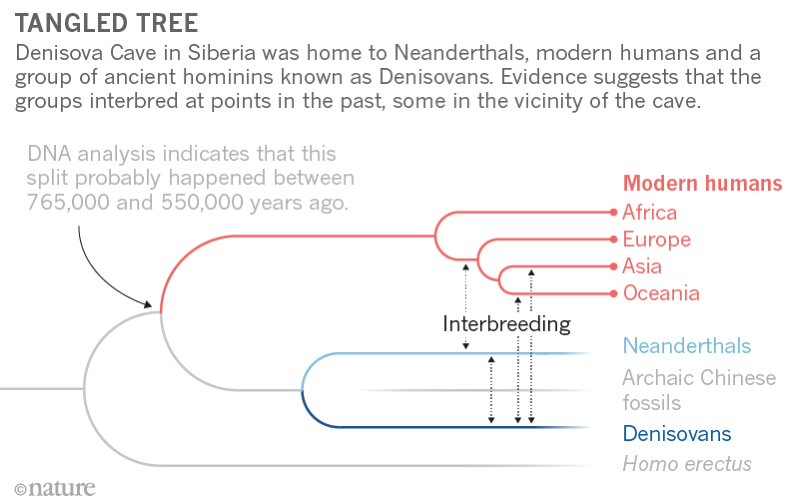 Viola, of the University of Toronto, and his colleagues compared a digital reconstruction of the skull fragment with corresponding parts of 112 present-day human skulls and 30 Stone Age Homo skulls, including Homo sapiens and Neandertals. The Denisovan find didn’t fit neatly into any previously known Homo species. Some features linked the Denisovan fossil to Neandertals and to a 430,000-year-old Spanish Homo species that had Denisovan ancestry (SN: 12/28/13, p. 8). The Denisovan skull fragment is surprisingly thick, more like cranial bones of Stone Age Homo erectus, Viola said. Such evidence is tough to interpret at this point, paleoanthropologist María Martinón-Torres of University College London said at the meeting. Interbreeding of closely related populations, such as Denisovans, Neandertals and H. sapiens, generates novel skeletal features that can obscure what started out as, say, a distinctive Denisovan look, she suggested. People on Papua New Guinea interbred with one genetically distinct Denisovan population around 46,000 years ago, the scientists estimate. Interbreeding with a second line of Denisovans took place by about 30,000 years ago and possibly as recently as 15,000 years ago. If the latter estimate proves correct in further studies, “Denisovans were the last surviving hominids who were not Homo sapiens,” Cox said. Those last survivors likely inhabited Papua New Guinea or a nearby island, he added.  The team probed about 3,000 DNA samples, obtained from around 100 communities on Papua New Guinea and other Southeast Asian islands, for molecular signs of interbreeding with Denisovans. The new DNA analyses suggest that some now living on Papua New Guinea and nearby islands carry roughly 400 Denisovan genes involved in immune and dietary functions. Denisovan DNA previously identified in present-day Siberians, East Asians and Native Americans does not occur in island Southeast Asia and thus represents a third Denisovan line, the researchers found. “We had no idea there were different Denisovan populations on Southeast Asian islands until now,” paleoanthropologist John Hawks of the University of Wisconsin–Madison said at the meeting. Further Denisovan populations may emerge from ongoing DNA studies of people in the Philippines, he predicted. |
|
|
|
Post by Admin on Sept 5, 2019 18:21:19 GMT
Abstract A fully sequenced high-quality genome has revealed in 2010 the existence of a human population in Asia, the Denisovans, related to and contemporaneous with Neanderthals. Only five skeletal remains are known from Denisovans, mostly molars; the proximal fragment of a fifth finger phalanx used to generate the genome, however, was too incomplete to yield useful morphological information. Here, we demonstrate through ancient DNA analysis that a distal fragment of a fifth finger phalanx from the Denisova Cave is the larger, missing part of this phalanx. Our morphometric analysis shows that its dimensions and shape are within the variability of Homo sapiens and distinct from the Neanderthal fifth finger phalanges. Thus, unlike Denisovan molars, which display archaic characteristics not found in modern humans, the only morphologically informative Denisovan postcranial bone identified to date is suggested here to be plesiomorphic and shared between Denisovans and modern humans.  INTRODUCTION In 2010, a small fragment of a finger phalanx recovered from the Denisova Cave (Denisova 3) in southern Siberia yielded a mitochondrial and a draft genomic sequence that changed our view of the evolution of the Late Pleistocene hominin lineages in Eurasia (1, 2), revealing a previously unknown archaic human population. The phylogenetic analysis of the Denisova 3 mitogenome yielded a divergence date from the ancestors of Homo sapiens and Neanderthals of around 1 million years (Ma) ago (1.3 to 0.7 Ma ago) (1, 3, 4), i.e., much earlier than the mitogenomes of the Neanderthals from the Late Pleistocene that diverged about 500 thousand years (ka) ago [690 to 350 ka ago; (3)] (Fig. 1). The nuclear genome, however, suggests a much more recent common ancestor between European Neanderthals (Vindija) and Denisovans dating to around 400 ka ago [440 to 390 ka ago; (5)], characterizing Denisovans as a sister group to Neanderthals (3, 5–8) (Fig. 1). Later, traces of an even more archaic human have been identified in the Denisova 3 nuclear genome (7), and a mitochondrial sequence related to that of Denisova 3 has been found in a ca. 400,000-year-old specimen from Sima de los Huesos (Spain), the nuclear genome of which is more closely related to Neanderthals than to Denisovans (3, 9). Together, these data suggest that the Denisovan mitogenome was either replaced with that of a more archaic human following an admixture event or represents the mitogenome of the common ancestors of Neanderthals and Denisovans before its replacement in the lineage of the Late Pleistocene Neanderthals (Fig. 1) (2–4, 9–10). The mitogenome of the late Neanderthals either could result from an introgression (i.e., replacement of the mitogenome following admixture) from early anatomically modern humans (AMHs) early after the separation of the AMH and Neanderthal populations, as proposed in one study (4), or could be due to incomplete lineage sorting given the uncertainties in the methods to estimate the dates and the wide confidence intervals of the dates proposed (Fig. 1). Furthermore, the comparison of the Denisova 3 nuclear genome with the genomic sequence of a roughly 100,000-year-old Neanderthal from the Denisova Cave revealed that Denisovans had also experienced gene flow from a Neanderthal population (Fig. 1) (7). Recently, a bone fragment, also from the Denisova Cave, has been found, through genomic analysis, to belong to a female individual that was the F1 hybrid of a Neanderthal mother and a Denisovan father (11). Her maternal Neanderthal contribution is more closely related to the genome of the 40,000-year-old European Neanderthal from Vindija (5) than to that of the ~100,000-year-old Neanderthal from the Denisova Cave. Furthermore, the paternal Denisovan genome of the hybrid appears to bear traces of an ancient Neanderthal admixture (11). These data indicate that gene flow between Neanderthals and Denisovans was not a rare occurrence.  Fig. 1 Model of the phylogeny of Neanderthal, Denisovan, and AMH populations over the past 1,400,000 years as deduced from both nuclear (blue envelope) and mitochondrial genomes (red lines). The vertical axis represents time in thousands of years (ka) ago. Population divergence dates estimated from genomic data and mitochondrial genome bifurcation date estimations originate from Prüfer et al. and Meyer et al., respectively (3, 5). Markers on the left indicate the means of the estimates for dates, and error bars indicate 95% confidence intervals. Gene flow events inferred from genome sequences are represented as dotted blue arrows (see text). Molecular dating methods based on mitochondrial sequences indicate that Denisovans must have inhabited the Altai region for over tens of thousands of years (12, 13). Despite the fact that all Denisovan mitochondrial sequences come from the same archeological site, Denisova Cave, the mitochondrial diversity of Denisovans is higher than that of Neanderthals spanning from Spain to the Caucasus (12). As inferred from the high-coverage Denisova 3 genome, the Altai population of Denisovans is characterized by low nuclear genome diversity, consistent with a prolonged small population size (10). Neanderthal populations also appeared to have been small, as assessed through the analysis of both the Altai and the Vindija genomes (5, 7). On the basis of the modeling, it has been proposed that despite reduced nuclear diversity of the individual local populations, the overall nuclear diversity of the Neanderthal metapopulation was higher (8), although this point remains under discussion as it varies with the modeling methods (6, 14). The extent of the Denisovan metapopulation diversity is still awaiting genomic characterization of remains originating from beyond the Denisova Cave, but the presence of Denisovan ancestry in modern human genomes suggests that there were at least two distinct Denisovan populations (15). Indeed, the comparison of the genomic sequence of Denisova 3 with the genomes of present-day humans has revealed interbreeding between Denisovans and early AMHs ancestral to present-day human populations not only in Southeast Asia, above all in Melanesians, but also in mainland East Asia (e.g., 15–18). The Denisovan ancestry in Melanesians appears to originate from a Denisovan population distantly related to that of the Denisova 3 specimen, and a similar ancestry can also be found in East Asia, particularly in Chinese and Japanese (15). In East Asians, a second Denisovan introgression from a Denisovan population more closely related to the Denisova 3 specimen was also detected (15). In some cases, the introgression proved to be adaptive, for example, in Tibetans (19) and Inuits (20). The distribution and diversity of Denisovan DNA in present-day human populations suggest that Denisovans were once widely distributed throughout Asia (15, 18). This evidence stands in contrast to the scarcity of unambiguously identified remains and of associated characteristic morphological features. What little morphological information that is available comes from a mandible from Xiahe on the Tibetan Plateau and three teeth from the Denisova Cave (2, 12, 13, 21). Denisovan mitochondrial genome sequences and low amounts of nuclear DNA have been recovered from a deciduous molar (Denisova 2) and two large-sized permanent molars (Denisova 4 and 8) (1, 12, 13), while the mandible has been identified as Denisovan based on proteomic information (21). The morphology of the Xiahe mandible is similar to that of the Middle Pleistocene specimens, such as the Chinese Lantian and Zhoukoudian, with features of the dental arcade shape that separate it from Homo erectus (21). It harbors some traits reminiscent of Neanderthals, while other Neanderthal-specific features are lacking (21). Thus, the rare Denisovan human remains identified to date show affinity to Middle Pleistocene hominins (2, 12, 13), particularly to those from China (21) and, to a lesser extent, to the Neanderthal lineage (12). The permanent molars from the Denisova Cave show complex occlusal morphology (1, 12, 13). Whether these peculiar characteristics of the molars are the consequence of introgression from a more archaic Eurasian population remains to be seen but cannot be excluded since such a low-level introgression has been identified in the Denisova 3 genome (7). Despite the importance of the Denisovan population for the study of human evolution, identification of Denisovan postcranial remains relies presently only on genomic data, since these remains of Denisovans exhibiting diagnostic features have yet to be reported. Progress in the identification of Denisovan skeletal remains would be instrumental for our understanding of this human lineage, for the identification of Denisovan remains, and for our ability to better characterize Denisovan population genomic diversity. Here, we report the morphometric analysis of a phalanx fragment that we show through its mitochondrial sequence to be the larger distal part of the original Denisova 3 phalanx, the genome of which had been published in 2010 and 2012 (1, 2, 10). In 2009, the phalanx was cut into two parts. The pictures of the phalanx taken by the Russian scientific team prior to its cutting, however, have been lost. The smaller proximal part of the bone was sent to the Max Planck Institute (MPI) for Evolutionary Anthropology in Leipzig, Germany, and sampling for paleogenomic analysis was performed. The larger distal part was sent to the University of Berkeley, CA, USA, and, in 2010, from there to the “Institut Jacques Monod” (IJM) in Paris, France, where it was measured and photographed and analyzed genetically. It was then returned to the University of Berkeley in 2011. The present analysis of both phalanx parts represents the first morphological study of nondental remains of this mysterious population that has inhabited Asia for hundreds of thousands of years, has interbred sometimes with Neanderthals and possibly with more archaic Eurasian humans, and continues to endure in the genomes of some present-day human populations. |
|
|
|
Post by Admin on Sept 6, 2019 18:02:37 GMT
RESULTS AND DISCUSSION Complete mitogenome sequence of the Denisova 3 fifth distal manual phalanx allows unambiguous matching of the two parts of the Denisovan phalanx The Denisova 3 phalanx (2008 Д-2/ 91) was identified in 2008 in layer 11.2 of the East Gallery, square D2, of the Denisova Cave, the date of which is assumed to be more than 50 ka ago (1, 2). To unambiguously match this distal phalanx fragment to the previously described proximal fragment, we retrieved its complete mitogenome [mitochondrial DNA (mtDNA)] sequence using DNA extraction and sequence capture procedures previously described (22–24). In total, 5838 unique reads were recovered, yielding a 26.7-fold coverage of the mitochondrial genome, of which each base was covered a minimum of twice (42 bases at twofold coverage) and a maximum of 70 times. The resulting consensus was identical to the previously published sequence (1) and represents the first replication of the Denisova mtDNA sequence outside of the MPI for Evolutionary Anthropology. This analysis also identified the previously proposed variable length in vivo of the polycytosine run at rCRS (revised Cambridge Reference Sequence) position 5889 (1), here found to be between 9 and 14 residues in length. This sequence identity indicates that the two phalanx fragments belong to the same individual. The exceptional preservation of endogenous DNA in the Denisovan phalanx is evident not only in its high endogenous DNA content but also in the length of the DNA fragments recovered. While an endogenous DNA content of 70% was recovered from the proximal fragment using the more sensitive single-stranded DNA library preparation method (10, 22), shotgun sequencing of the distal fragment analyzed in this study prepared using a double-stranded library method contained 11.3% endogenous content. This is in line with the ~6-fold increase in endogenous content reported between these two methods (10, 22). Previous analyses of the distribution of the endogenous DNA fragment lengths were performed using only merged reads, which prohibits the identification of endogenous molecules longer than 134 nucleotides. The paired-end mapping strategy used in this study reveals a more complete endogenous fragment length distribution. We show all Denisova-mapping DNA fragments to have a mean distribution of 86.7 base pairs (bp), with a median of 81.3 (Fig. 2). The mean value is similar to that first reported [mean, 85.3 bp; (1)], although these two means are skewed toward larger fragments since the library construction method used for these two studies did not recover the greater part of shorter fragments found in libraries prepared with the single-stranded library method (10). The longest fragment containing diagnostic Denisovan nucleotide sites recovered in this study was 236 bp.  Fig. 2 Distribution of DNA fragment lengths mapping to the Denisova mitochondrial sequence. Fragment lengths given in 10-bp bins are shown. Median is indicated by the dotted orange line. Read pairs that did not overlap sufficiently to be merged (i.e., longer than 134 nucleotides) but that could be mapped as paired-end reads were analyzed individually, and fragments carrying Denisovan single nucleotide polymorphisms (SNPs) were kept. In contrast to the analyses of the proximal half of the phalanx, which reported low levels of modern human DNA contamination (0.35%) (1), the distal half showed the presence of a much higher modern human mitochondrial contaminant (12.1% detected by a similar method). Further investigation revealed that this contaminant could be attributed to a single haplogroup, J1b1a1. Since no member of the IJM laboratory in Paris where the genetic analysis was performed carried mitochondrial haplogroup J, we suspect this contamination to have occurred at some point during the previous handling of the sample, prior to its preparation for genetic analysis. Anatomical description When the mitogenome analysis of the proximal epiphysis from the metaphyseal surface indicated that the specimen was not a recent modern human, it was digitized through microcomputed tomography (μCT) at the Department of Human Evolution at the MPI for Evolutionary Anthropology, Leipzig (courtesy of H. Temming and J.-J. Hublin). The reconstructed image based on these scans is shown in Fig. 3. Further sampling for nuclear DNA analysis was performed on this specimen (2), and two small holes on the articular surface witness the drilling procedure. The proximal fragment of the phalanx is now composed of two pieces (the proximal epiphysis and the remains of the dorsal part of the diaphysis) (Fig. 3).  Fig. 3 Views of the Denisova 3 DP5. (A) Virtual reconstruction of the Denisova 3 DP5 in dorsal view. Three fragments of the Denisova 3 DP5 are shown. Natural color: Photograph of the distal two-thirds of the phalanx; green: semiring of the dorsal surface of the proximal extremity of the diaphysis of the reconstituted image based on the μCT scan; blue: its proximal articular surface. (B) Virtual reconstruction based on the μCT scan of the Denisova 3 DP5 in palmar view. (C) Virtual reconstruction of the proximal articular fragment in distal (top) and lateral (middle) views, and the dorsopalmar μCT section of this piece at the level of the fusion (bottom). The D-P line (D, dorsal; P, palmar) represents the location of the section on the distal view and helps to orient the bone in the lateral view. The red arrows indicate the fusing zone. (D) Additional views of the distal fragment and the virtual reconstruction of the proximal part of the Denisova phalanx. 1 and 2: Lateral views of the distal fragment; 3: distal; 4: proximal; 5 and 6: lateral views of the proximal fragment. The gray surfaces on the outer border of the phalanx correspond to the forceps with which the phalanx was held while the photo was taken. Photos of the distal part of the phalanx were taken by E.-M.G., IJM, CNRS, Université de Paris, UMR 7592, Paris, France. The renderings of the μCT scans and the virtual reconstruction were performed by B.V., Department of Anthropology, University of Toronto, Toronto, Canada. The larger distal part of the Denisova 3 phalanx was documented in Paris by measurements with a high-precision vernier caliper (Table 1) and high-resolution images under a stereomicroscope (Leica MZ FLIII, PLANAPO 0.63×) prior to sampling (Fig. 3, A, B, and D). The reconstituted image of the entire phalanx is shown in Fig. 3 (A and B) as a virtual reconstruction in the dorsal (Fig. 3A) and palmar (Fig. 3B) views of the distal and proximal part of the phalanx, combining the photographs of the distal part and the three-dimensional (3D) model of the proximal parts. Lateral views of the distal part and several additional views of the proximal part are also shown (Fig. 3D)
Table 1 Measurements of the distal part of the DP5 of Denisova 3.
Measurements (in mm) of the distal part of the Denisova 3 phalanx [Denisova Cave 2008/East Gallery/layer 11.2/square D2 Phalanx tertia (probably V)] were taken with a vernier caliper directly on the bone fragment and on rectified photographs where applicable.
Measurement With caliper on
distal part (mm) On rectified
photographs (mm) Comments
Midshaft breadth 3.60 3.7
Proximal breadth 5.62 ? Cut basis
Distal breadth 4.52 4.65
Maximum length 11.28 Basis cut in a nonperpendicular way
Distal height 2.80
Midshaft height 3.60 (Difficult since there is no "promontory"
above which the measurement should be taken;
here: maximal width)
Here, we reanalyzed the μCT scans and photographs of the proximal fragments (the articular surface and the semiring representing the dorsal half of the proximal end of the diaphysis), as well as the photographs of the distal fragment in comparison with the distal phalanges (DPs) of Neanderthals and of Pleistocene and recent modern humans at various stages of development (table S1; see below). The distal border on this surface shows traces of the sawing of the phalanx into two pieces, consistent with those observed on the distal fragment and the proximal semiring fragment of the diaphysis. We draw the conclusion that the morphology of Denisova 3 is incompatible with an unfused immature distal phalanx. First, the palmar surface of the apical tuft is characterized by a well-defined ungual tuberosity, as is the proximal V-shaped ridge of the tuft for the insertion of the flexor digitorum profundus tendon sheath (Fig. 3B). Second, the proximal articular surface fragment on the palmar surface exhibits a relief in the middle part that is unlike the morphology of an unfused proximal epiphysis (Fig. 3B). Last, the μCT images confirm that the distal part on the palmar surface of the articular fragment is a piece of the diaphysis that is fusing (Fig. 3C, red arrows), not the original border of the proximal epiphysis. Both the semiannular dorsal surface of the diaphysis and the dorsal part of the proximal articular fragment present rounded borders that are consistent with the epiphysis fusing at the time of death (Fig. 3A). Since it takes between 2 and 4 months for an epiphysis to complete the process of fusion, once started, we conclude that the dimensions of the phalanx are close to its final mature state (25). In summary, the evidence from both the distal and proximal fragments indicates that the Denisova 3 DP5 (fifth distal phalanx) belonged to an adolescent. Nuclear DNA analyses show that this individual was a female, allowing us to narrow the age at death around 13.5 years based on the standards from extant humans and assuming that Denisovans had a fairly similar development. If we accept that the phalanx is close to the mature state, then it is possible to tentatively identify both digit and side for Denisova 3. Considering extant modern human diversity, the estimated maximum length of Denisova 3 falls best within the variability of the DP5s (25). In addition, the asymmetry of the ungual tuberosity and the curvature of the shaft in the dorsal view indicate that this DP5 is likely from the right side [(26); I. Crevecoeur, personal observation]. |
|
|
|
Post by Admin on Sept 6, 2019 18:35:43 GMT
Morphometric comparison We performed morphometric analyses of the DP5 of Denisova 3 based on the measurements taken on the original specimen and the virtual reconstruction for the maximum length (see Fig. 4A for a schematic representation of the various measurements considered here). Using univariate and multivariate analyses, we compared these measurements with data from published and unpublished DPs of Neanderthals, Pleistocene modern humans, and three samples of recent modern humans from France and Belgium dated from the Neolithic to the Middle Ages (table S1; see Fig. 4B for a direct side-by-side comparison of a DP5 from a Neanderthal, a Denisovan, and an AMH). The dimensions of Denisova 3 and the means and SDs of the comparative samples are given in Table 2. The comparison sample includes one AMH and one Neanderthal specimen in which the proximal epiphyses were in the process of fusing.  Fig. 4 Measurements of the Denisovan fifth finger phalanx and comparison with those of Neanderthals and AMHs. (A) Schematic representation of the various measurements of digital phalanges reported in Table 2 and here: ML, maximal length; PH, proximal height; PAH, proximal articular height; PB, proximal breadth; PAB, proximal articular breadth; MH, midshaft height; MB, midshaft breadth; DB, distal breadth; DH, distal height. (B) Comparison of the dorsal view of the DP5 of a Neanderthal (Krapina 206.12), a recent modern human, and the reconstructed Denisova 3. (C) Scaled Z-scores of the Denisova 3 dimensions (Den) compared to both Neanderthal (NEAND) and pooled modern human (MH) ranges of variation. Den_Neand, Den_RMH, Den_NDP5, and Den_MDP5 indicate the comparison of the values of Denisova’s DP5 with the mean and SD of each comparative group from Table 2: NEAND_DP and MH_DP, all distal phalanges; NDP5 and MDP5, DP5s only. Z-scores were scaled in a way that zero represents the mean of each range of variation, and +1/−1 represent the upper and lower 95% limits of each range of variation, respectively (see Materials and Methods). Therefore, when a value is lower than −1, it means that it falls outside the lower statistical limit (P = 0.05) of the range of variation of the comparative group in question. (D) Bivariate plot of the two first principal components of the PCA on size-adjusted measurements of the DPs. MH_DP, pooled sample of Pleistocene and recent modern human DPs; NEAND_DP, Neanderthal DPs. MH-DP5f, fusing DP5 from the modern human sample. (E) Correlation circle between the measurements of the distal phalanx and the two first principal components of the PCA on size-adjusted measurements of the DPs shown in (D). Photos of the distal part of the phalanx were taken by E.-M.G., IJM, CNRS, Université de Paris, UMR 7592, Paris, France. The renderings of the μCT scans and the virtual reconstruction were performed by B.V., Department of Anthropology, University of Toronto, Toronto, Canada. Photos of the Krapina 206.12 specimen from the Croatian Natural History Museum collections, Zagreb, Croatia, and from the recent human specimen from the collections of UMR 5199 PACEA, Université de Bordeaux, France, were taken by I.C., UMR 5199 PACEA, Université de Bordeaux, France. With the possible exception of the proximal breadth, all dimensions of Denisova 3 fall within the range of variation of modern human DP5s (Fig. 4C). The dimensions of the proximal extremity fall in the lower part of the modern human DP5 variation and outside that of the Neanderthals with regard to the articular surface measurements. This may be related to the state of preservation of the proximal extremity and, probably, also to its state of fusion. While the midshaft height of the diaphysis and the distal height of the apical tuft are close to the modern human mean, the remaining measurements fall into the lower range of the variation, indicating that the Denisova 3 phalanx is gracile. Nevertheless, the fact that we are dealing with an adolescent female must be taken into consideration with regard to potential size and gracility. We performed a multivariate analysis using size-adjusted dimensions to allow a comparison of the DPs based on shape rather than size. The projections along the two first principal components are given in the two following bivariate plots. The scatterplot of the individuals is illustrated in Fig. 4D, and the correlation circle for the original variables in Fig. 4E. The first two principal components represent more than 50% of the total variation. A clear distinction is visible between the Neanderthal and the modern human samples, with Denisova 3 positioned in the lower right quadrant within the modern human variation (Fig. 4D). As expressed by the correlation circle, the Denisovan DP5 differs from that of the Neanderthals in that the former combines a narrow apical tuft (distal breadth) with a thicker DP, particularly at the midshaft and proximal end (midshaft height and proximal height) (Fig. 4E). Neanderthal DPs have been usually described as notably different from modern humans because of their length and the shape and dimensions of their apical tufts [e.g., (27, 28)]. Neanderthal DPs are proportionally longer with wider extremities compared with modern humans, which gives the impression of flattening of the bone (29). This conformation of the apical tuft among Neanderthals seems to be related to functional rather than cold climate adaptations (30). These characteristics are confirmed by our analysis, but additional observations can be made regarding the DP5s. Neanderthal DP5s seem to occupy a specific position compared with the other Neanderthal DPs. This difference between the fifth and the other phalanges that is not visible in the modern human sample is due to both the specific morphology of the Neanderthal DP5 compared with the other digits, driven by the shape of the midshaft and the apical tuft, which are both narrower than the remaining digits. On the contrary, when not taking into account the size factor, AMH DP5s scatter within the variability of the other DPs. CONCLUSIONS We could genetically link the distal part of a DP5 from the Denisova Cave in Siberia to the Denisova 3 phalanx fragment, whose genome identified it as a representative of a population more closely related to Neanderthals than to modern humans. Morphometric analysis based on high-resolution pictures, linear measurements, and the comparison with the DP5s of Neanderthals as well as Pleistocene and recent modern humans shows that it is within the range of variation of the dimensions of the DP5s of modern humans and distinct from that of Neanderthals. We propose that this represents the plesiomorphic morphology within the genus Homo (32), consistent with the morphology of early Homo DPs (31, 32), and that the derived morphology of the Neanderthal phalanx evolved after their split from the ancestors of the young woman from the Denisova Cave (see Fig. 1). This finding calls for caution when identifying potential Denisovan postcranial skeletal remains beyond Denisova, as their morphology might be ambiguous or more similar to modern humans than to Neanderthals. Science Advances 04 Sep 2019: Vol. 5, no. 9, eaaw3950 |
|
|
|
Post by Admin on Jan 7, 2020 20:29:39 GMT
Since 2008, when their fragmentary fossils first turned up in a Siberian cave, the Denisovans have been the most mysterious branch of the human family tree. The archaic humans, like Neanderthals, lived at the same time as early Homo sapiens but have been extinct for tens of thousands of years. 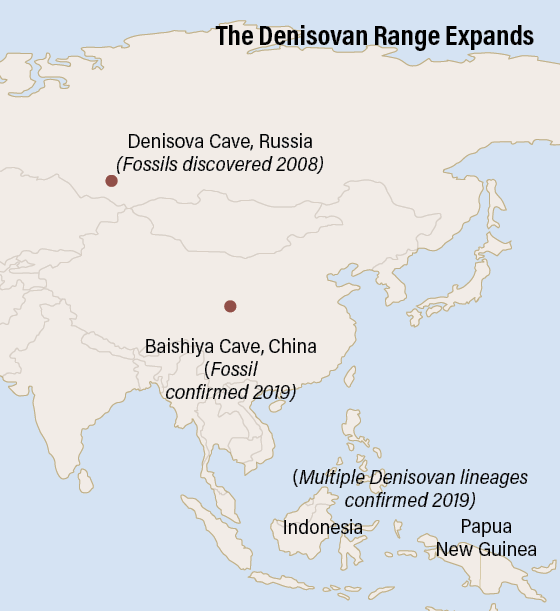 For nearly a decade, we’ve known Denisovans from only one site, which yielded just a shard of finger bone and a few teeth from four individuals — and ancient DNA extracted from the scant remains. In 2019, however, a series of discoveries revealed our evolutionary kin in greater detail than ever, answering some questions but also presenting new mysteries. One thing is clear: The Denisovans were much more complex than we thought, occupying diverse and sometimes extreme environments spread over much of Asia. In March, researchers announced that they had found two small skull fragments — the first known pieces of a Denisovan braincase — belonging to a fifth individual in Denisova Cave, in the Altai Mountains of southern Siberia. However, the biggest news about our mysterious extinct cousins came from outside their namesake cave. |
|














