|
|
Post by Admin on Nov 12, 2014 14:32:02 GMT
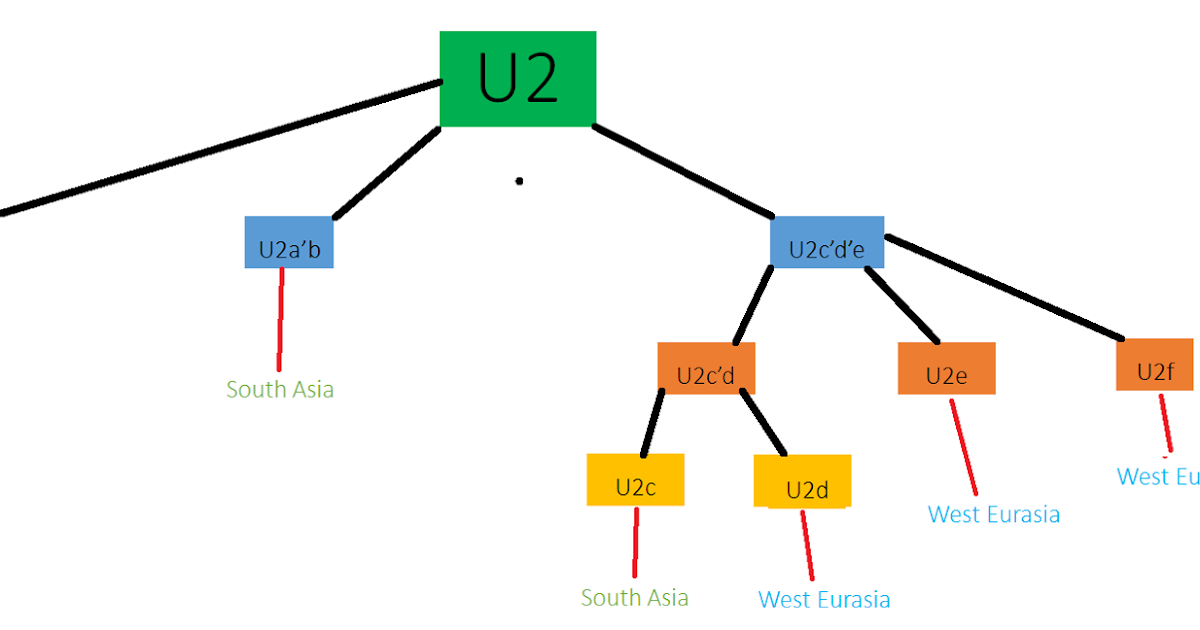 Haplogroup U2 is rare lineage very homogeneously spread across most of Central Asia, Europe, the Middle East and North Africa, with a frequency typically ranging from 0.5% to 2%. Only a few isolated ethnic groups, mostly in the Volga-Ural and North Caucasus regions, have frequencies above 3%. This includes the Uralic-speaking Udmurts (10%) and Mordvins (7%), as well as the Karachay-Balkars (4.5%), Nogays (3.8%), North Ossetians (3.6%), Adyghe-Kabardin (3.6%) and Dargins (3.6%) in the North Caucasus, and the Latvians (3.5%) in the East Baltic. The only region where U2 is constantly found in higher frequencies is South Asia, where it is found found in roughly 6.5% of Bangladeshi people, 12% of Sri Lankans, and at an average frequency of 5.5% of in India, especially among Indo-Euopean speakers (7.5%) and with local peaks in northern India exceeding 20% (source: Mestpalu et al. 2004). However, South Asian subclades of U2, namely U2a, U2b and U2c, differ from the Central Asian U2d and European U2e. Only a few ethnic groups appear to completely lack haplogroup U2, although this could be due to sampling bias. So far, U2 has not been found among Ashkenazi Jews, Cypriots, Sardinians, Welsh, Icelandic, Saami, Lithuanians, Avars and Chuvash people. Haplogroup U2 is an extremely old lineage, going back at least 40,000 years, when Homo sapiens first expanded from the Middle East into South Asia and Central Asia, and before they even set foot in Europe. The oldest Homo sapiens DNA sample from Europe tested to date, a 33,000-year old Cro-Magnon from the Kostenki site on the Don River in the Russia, belonged to haplogroup U2 (see Krause et al. 2010). Further U2 samples were identified among Mesolithic European hunter-gatherers, including a 11,000 year-old U2e from Blätterhöhle in Germany (Bollongino et al. 2013), two 9,500 year-old U2e individuals from Karelia in Russia (Der Sarkissian 2011), and two 8,000 year-old U2e1 individuals from Motala in Sweden (Lazaridis et al. 2014). Based on these ancient DNA results from Europe and the presence of all basal subclades of U2 in Central Asia, it is likely that U2 people roamed between Central Europe and Central Asia during the Paleolithic and Mesolithic, and perhaps already in other parts of Europe and in South Asia. The steppes of eastern Europe and Central Asia are probably the original geographic location from which such a dispersal was made possible during the Stone Age, and again during the Bronze Age. U2 became much scarcer among European Neolithic samples, only popping up once in an early Linear Pottery sample from Hungary. In the late Copper and early Bronze ages, U2 made a come back among Proto-Indo-Europeans cultures. U2 samples were found in the Yamna culture (U2e1a), Corded Ware culture (U2e1 and U2e2), Unetice culture (U2e1f), as well as the Andronovo culture (U2e) in Central Asia. Proto-Indo-European speakers from eastern Europe had a higher proportion of Mesolithic European ancestry than Neolithic farmers, so it isn't surprising to find a slightly higher frequency of U2e among samples from that period. U2e actually shows up with surprising regularity in ancient samples from Ukraine and European Russia. For example it was also found in Iron Age Scythian remains from Rostov-on-Don in southern Russia. U2e even showed up in Indo-European bones from the Tarim basin in north-west China, also dating from the Iron Age (possibly Scythian or Tocharian), but also at a Xiongnu (Hunnic) site from the same period in Mongolia. Haplogroup U4 is found at a frequency ranging from 2% to 6% in most regions of Europe. Its highest frequency is observed among the Chuvash (16.5%), Bashkirs (15%) and Tatars (7%) of the Volga-Ural region of Russia, followed by Latvia (8.5%), Georgia (8.5%), Serbia (7%), and southern Daghestan (6.5%). Generally speaking, U4 is more common in Baltic and Slavic countries and around the Caucasus than anywhere else. Within Europe U4 is rarest in fringe regions such as Ireland (1.5%), Portugal (1.5%), north-west Spain (0.5%, except Cantabria which has 3%), Finland (1%), and especially among the Welsh, Sardinians and Saami, where it is completely absent. U4 is not found in countries or regions that lack the paternal lineage R1a, with which it seems to be intimately linked. Outside Europe and the Caucasus, U4 is found especially in Iran (3%) and throughout Central Asia, particularly in Kyrgyzstan (3%), Turkmenistan (3%), Uzbekistan (2.5%) and Kazakhstan (2%), but also in parts of Siberia, notably in the Altai Republic (5%) and among the speakers of the Khanty and Mansi languages (12%), east of the Ural mountains. U4 is also found at high frequencies in some ethnic groups in Pakistan and Afghanistan, including among the Balochi (2.5%), Hunza Burusho (4.5%), Hazaras (8%), Parsi (13.5%) and especially among the Kalash (34% according to Quintana-Murci et al. 2004), although these frequencies have to been taken cautiously as they are based on very small sample sizes.  Haplogroup U4 is found at the highest frequency in the Indo-Aryan peoples of Pakistan and the Kalash people (34%). Haplogroup U4 originated approximately 25,000 years ago, during the Last Glacial Maximum (LGM). U4 appears to have been a relatively common lineage among Mesolithic European hunter-gatherers. It was identified in skeletons from Mesolithic Russia (including some U4a1 samples), Lithuania, Sweden, Germany and Portugal. Based on the small number of Mesolithic samples tested to date, U4 seems to have been much more common in Northeast Europe than elsewhere. This would make sense since it correlates strongly with Y-haplogroup R1a nowadays. During the Neolithic period U4 stands out by its absence from the hundreds of samples tested to date, except for one Late Neolithic/Chalcolithic sample (c. 3250 BCE) from Catalonia and one from Portugal (3000 BCE). Along with Cantabria, Catalonia and Portugal also happen to be the regions of Iberia where U4 is the most common today. As there appears to be a continuity in these regions since the Mesolithic, it is possible that Iberian U4, or West European U4 in general, was brought by nomadic tribes of hunter-gatherers belonging to old, pre-Indo-European subclades of R1a, such as SRY1532.2 or M17. Originally from eastern Europe, these R1a/U4 people would have crossed all Europe and survived in isolated pockets of western Europe from the Neolithic onwards. Haplogroup U4 make a strong come back during the Bronze Age, where it is found at high frequency among remains from the Proto-Indo-European Corded Ware culture and Catacomb culture (a staggering 25% of the 28 samples, see Wilde et al. 2014)), both associated with the diffusion of R1a to Central Europe and Scandinavia, and in the Unetice culture, thought to be the first predominantly R1b culture around what is now Germany. The subclades identified for the Corded Ware and Unetice cultures were respectively U4a1 and U4c1. Both of these subclades are also found in Central Asia today, confirming the Indo-European connection. U4 was also found in the Yamna culture, the presumed homeland of Proto-Indo-European speakers in the Pontic Steppe. The Volga-Ural region played a major role in Bronze Age PIE cultures, and remained fairly isolated from the subsequent population movements within Europe. The same is true for the central Caucasus region, such as Georgia and southern Daghestan, which received relatively little influx of foreign genes after the Bronze Age. The fact that U4 is many times more prevalent in these regions today also suggest a higher frequency among Bronze Age PIE speakers. During the Yamna and Maykop periods (3700-2500 BCE), R1a and R1b people would have intermingled in Pontic-Caspian Steppes and North Caucasus, explaining why U4 is also found among R1b populations, although at a lower frequency than among R1a populations. In modern France and northern Italy, the percentage of U4 looks directly proportional to the frequency of combined haplogroups R1a and R1b. Interesting, Fernández et al. (2005) also found two U4 individuals (including one U4a2b) in Sumerian city of Mari in Syria dating from the Early Dynastic Period (2900-2700 BCE), just after the Uruk collapse, which could have been caused by early Indo-European incursions into the Near East. U4 maternal lineages were found in Bronze Age cultures associated with the Indo-European migrations in Central Asia and Siberia, such as the Andronovo and Karasuk cultures (Keyser 2009), but also in in the Tarim basin in north-west China during the Early Iron Age (Zhang 2010). |
|
|
|
Post by Admin on May 30, 2016 22:51:40 GMT
 A study by Arredi.et al (2004) includes the frequencies of lineages among one Kabyle population from Tizi Ouzou province. MtDNA Haplogroups, inherited only from the mother, were found at the following frequencies: H (32.23%), found throughout Europe; U* (29.03% with 17.74% U6), common to North Africa; preHV (3.23%), preV (4.84%), V (4.84%), T* (3.23%), J* (3.23%), L1 (3.23%), L3e (4.84%), X (3.23%), M1 (3.23%), N (1.61%) and R (3.23%). After the dispersal of modern humans Out of Africa, around 50–70 ky cal BP1,2,3,4 or earlier based on fossil evidence5, hominins with similar morphology to present-day humans appeared in the Western Eurasian fossil record around 45–40 ky cal BP, initiating the demographic transition from ancient human occupation (Neandertals) to modern human (Homo sapiens) expansion on to the continent1. The first insights of the genetics of early Eurasian modern humans were recently provided by four ancient human genomes: Ust’-Ishim (Western Siberia, 45 ky cal BP)6, Kostenki (Russia, 39–36 ky cal BP)7, Fumane 2 (Italy, 41–39 ky cal BP)8 and Peştera cu Oase (Romania, 37–42 ky cal BP)9. Population genetic analyses of modern-day human mitochondrial haplogroup distributions suggest that in conjunction with the Eurasian expansion, some populations initiated a back-migration to North Africa10,11,12,13. Although the first genome of an ancient African individual (Ethiopia, 4.5 ky cal BP) identified a back-migration from Eurasia to Africa within the last 4.500 years14, the scarcity of older human remains in North Africa has prevented researchers from obtaining direct evidence of such a migratory phenomenon during the Paleolithic period. We present the mitochondrial genome (mitogenome) of the Peştera Muierii 1 (PM1) remains from Romania, directly dated to 35 ky cal BP15, which sheds new light on the Early Upper Paleolithic (EUP) migrations in Eurasia and North Africa. We estimated the phylogenetic position of PM1 using Bayesian inference in a two-step analysis. First, we aligned the reconstructed mtDNA sequence with 10 other ancient mitogenomes, including two Denisovans18, two Neandertals19 and 6 ancient Homo sapiens from the EUP2,6,7,9 (Fig. 1A and Supplementary Table 2). The tree fully supports the position of PM1 within the modern Homo sapiens clade (Fig. 1A). None of the 63 ‘diagnostic’ positions (at which ten Neandertal mitogenomes differ from 311 present-day humans) appeared in PM119,20,21,22,23,24. This observation is compelling as the morphology of PM1 exhibits features related both to modern humans and Neandertals15. Furthermore, the PM1 remains are not associated with any particular cultural techno-complex, as the lithic artifacts found at the site were related both to Mousterian (associated with Neandertals) as well as to Aurignacian assemblages (associated with early Homo sapiens)25. None of the reported mtDNA sequences from early modern humans have displayed Neandertal mitochondrial genomes4,6,7,8,9, although a low level of admixture has been detected in the nuclear DNA of modern humans24,26 and at higher proportions in one Paleolithic human9. As a second step, we estimated the mitogenomic position of PM1 within modern humans by analyzing 144 modern27 and 47 ancient human mitogenomes covering the known mitogenomic variability (Supplementary Tables 2 and 3). The haplogroup of PM1 falls within the U clade (Fig. 1B and Supplementary Table 3), which derived from the macro-haplogroup N possibly connected to the Out of Africa migration around 60–70 ky cal BP1,2,3,4. In line with this, the Peştera cu Oase individual that lived on the current territory of Romania, albeit slightly earlier than PM1 (37–42 ky cal BP) also displays haplogroup N9. Figure 1: Phylogenetic analyses of the Peştera Muierii-1 (PM1) mitogenome (35 Kcal BP, Romania).  (A) Unconstrained Bayesian phylogenetic analysis including ancient H. sapiens, Neandertals and Denisovans. (B) Unconstrained Bayesian phylogenetic analysis including ancient and present-day H. sapiens. The tree is time-calibrated using node ages. The color of node dots indicates the posterior probability (pp): green dots = maximum robustness, yellow dots = slight robustness, red dots = low robustness. The analysis of the PM1 mitogenome polymorphisms revealed 15 nucleotide changes with respect to the rCRS28, identifying the PM1 mitogenome as a basal haplogroup U6* (Supplementary Table 1). One of these polymorphisms is a private mutation, T10517A, not previously found in any mitochondrial genome. The U6 haplogroup is the only sub-haplogroup within the U clade currently present in Africa, showing an increasing frequency gradient from Eastern (1.09–1.57% in Egypt) to Western North Africa (8.89% in the Magreb). A similar longitudinal gradient is present in the Southern European populations (from 0.19% in Eastern Mediterranean to 1.12% in South Spain)29,30 (Fig. 2B). The U6 haplotypes found in present-day Europeans have been attributed to African sources, mainly to the historic Moorish expansion, but also to prehistoric influence since Neolithic times29,30. Hence, PM1 is the first basal U6 haplogroup found in Europe that is not connected to recent migration from Africa. The mitogenome from PM1 offers important information in order to understand human population movements during the Paleolithic Age related to the haplogroup U6. While all the extant U6 haplotypes belong to derived branches, i.e. U6a’b’d (characterized by transition, 16219) or to the less frequent U6c (characterized by a set of eleven mutations, 150, 437, 793, 3688, 4965, 5081, 11013, 13879, 15244, 16169, 16189)30 (Fig. 2A), the haplotype of the PM1 individual belongs to the basal U6 haplogroup from which the rest of haplotypes were derived (Fig. 2A). This scenario confirms that the U6 mitochondrial lineage has a Eurasian origin, supporting the hypothesis of an early back-migration from Eurasia to North Africa in the EUP10,11,30. Figure 2: Distribution of the U6 mitochondrial lineages.  Individuals carrying haplogroup U possibly spread westward from Western Asia around 39–52 ky, reaching Europe as signaled by haplogroup U5, and North Africa signaled by haplogroup U6, which likely represents a genetic signal of a EUP return of Homo sapiens from Eurasia to North Africa11,29,30. The time of the most recent common ancestor (TMRCA) for U6 was estimated to 35.3 (24.6–46.4) ky BP29,30. Thus it has been proposed that the lineage originated somewhere in Western Asia11,29,30. We found a basal U6 in South East Europe, on the current territory of Romania 35 ky BP, suggesting that either the U6 lineage originated in Eastern Europe or the TMRCA of U6 is older than 35 ky. Our estimates of the haplogroup U6 TMRCA that incorporate ancient genomes (including PM1) set the formation of the U6 lineage back to 49.6 ky BP (95% HPD: 42–58 ky) (using a mutation rate of 2.06* 10−8 SD = 1.94 * 10−9) (Fig. 1). Our estimates are almost identical in age to that by reference11 (45.1 ± 6.9 ky). Given the presence of a basal U6 mitogenome in Romania 35 ky BP, the distance between Western Asia and Romania, and the estimated diffusion pace of hunter-gatherer populations30 suggest that the early populations carrying haplogroup U6 most likely started their spread to Eastern Europe before 40 ky BP. It is unclear whether the haplogroup U6 diversified in Africa or arrived to the continent as an already diversified lineage. However, the detection in South East Europe (Romania) of a basal U6* haplotype presenting only two of the diagnostic mutations (3348 and 16172) of modern-day U6 haplogroups (Fig. 2A and Supplementary Table 3) strongly points to an “on route” differentiation of U undifferentiated lineages to basal U6 lineages before reaching Africa. Scientific Reports 6, Article number: 25501 (2016) |
|
|
|
Post by Admin on Jun 2, 2016 22:57:11 GMT
 The complete mitochondrial genome of the young man from Byrsa was identified as belonging to haplogroup U5b2c1 possessing all of the defining mutations with the exception of 16192T, one of the two mutations defining U5, which has shown to be unstable with frequent reversions [37]. He also had five additional, coding region mutations (Fig 4). None of the researchers who collected or processed the Phoenician sample have mitochondrial haplotypes belonging to Haplogroup U. All methods to avoid contamination were applied and the possibility of modern contamination in the sequence obtained was assessed using standard aDNA authenticity criteria and shown to be unlikely, thus we are confident that the haplotype identified is indeed that of the young man from Byrsa.  Given the reputed Lebanese origins of the founders of Carthage, we undertook full mitochondrial genome sequencing of 47 modern Lebanese samples that had previously been typed to Haplogroup U through analysis of the HVR-1 [12]. Haplogroups identified and haplogroup frequency are shown in Table 1. Only seven of the modern Lebanese samples belonged to Haplogroup U5 and of those, two were U5b, but neither belonged to U5b2c or derived haplotypes.  A phylogenetic analysis of all U5b2c sequences available in GenBank or from published supplementary data (Z2478) is shown in Fig 5. Haplogroup U5 is considered to be one of the most ancient haplogroups in Europe and is believed to have arisen there [43]. The coalescence time estimate for U5 is 29.6 kya (22.7–37.2 kya) [25] and 20–24 kya for U5b [44]. It is not uncommon in Mesolithic European populations, particularly those from Central and Eastern Europe [45]. Haplogroup U5b2c1 has been identified in both La Braña 1 and 2, the 7000 year-old remains recovered from the La Braña-Arintero site in León in Northwestern Spain [42]. Our Phoenician differed from the La Braña 1 complete mitochondrial genome at eight sites (positions 3882, 5351, 5773, 6023, 9869, 16069, 16126, and 16192). The mutations at sites 16069 and 16126 appear to be private mutations for La Braña. It appears that the U5b2c1 haplogroup is rare in modern populations, with only a few modern sequences published [33, 38, 46] or available in public databases (Family Tree DNA). All of the reported U5b2c1 carriers are of presumably (if not specifically stated) European ancestry, from Spain, Portugal, England, Ireland, Scotland, the United States and Germany. Three of the additional non-defining mutations found in our Phoenician, 5351G, 6023A, and 9869T, are shared with one “European” sample [46] and an individual from central Portugal [33]. Interestingly, Fig 5 shows that our Phoenician sample is most closely related to the modern sample from central Portugal. The unidentified “European” sequence (EF758625) which was deleted from the analysis due to missing data, also carries the three mutations which define the branch on which we find the Phoenician and the Portuguese sample. A separate branch within U5b2c1 contains five samples, from England and Germany or otherwise unidentified as to location of origin, with the La Braña Mesolithic sample (JX186998) on its own branch. Given the limited numbers of published full mitochondrial genomes, it is difficult to identify a likely origin for the mutations defining the Phoenician and Portuguese branch, but it is currently not inconsistent with a Southwest European origin. While the U5b2c2 sequences all originate in Central- and North-Western European or likely derived populations (e.g. US Caucasian), and the U5b2c sequences in GenBank (KC847158 and KU587510, which was excluded from the analysis due to missing data) originate in Germany and England respectively (both samples from Family Tree DNA), it is likely that U5b2c originated in Central/Northwestern Europe. Future complete mitochondrial genome sequencing of European populations, both ancient and modern, will undoubtedly provide more information regarding the distribution and frequency of the U5b2c1 haplogroup across the continent and thus will help us to reconstruct its evolutionary history. It is generally argued that Mesolithic populations carrying high frequencies of U5 were replaced by Neolithic populations expanding into Western Europe during the Neolithic transition [47]. Recent analyses of ancient DNA suggest that there were two waves of Neolithic expansions that dramatically influenced the genetic makeup of Western Europe [48–51]. An early Neolithic expansion out of Anatolia, occurring some 7000–9000 years ago, moved into Western Europe and replaced many Mesolithic Hunter-Gatherer populations, resulting in an increase of Near Eastern mitochondrial lineages. After this initial invasion of Neolithic farmers, there appears to have been a resurgence in the Western Hunter-Gatherer genetic signatures, followed by a second migration and replacement in many locations from the Eurasian Steppes, associated with the spread of the Yamnaya culture, beginning some 4500 years ago [49]. Several other more regionally focused aDNA studies, however, have shown that there are different patterns of population replacements in various regions of Europe [52, 53]. Unlike the early Neolithic arrival in the Iberian peninsula, which likely arrived in coastal regions [54, 55], it is possible that the second, Eurasian Steppe derived, replacement occurred later or had less of an impact on the southern and western Iberian coast and other Mediterranean coastal regions than in more northern or eastern inland locations [49, 53] and thus the Western Hunter-Gatherer mtDNA lineages, including U5b2c1, may have been more common there at the time of Phoenician contact and settlement. While the U5b frequency in modern populations in Western Europe is less than 2% it is much rarer in today’s Near Eastern populations [12]. In our full mitogenome sequencing results of 47 modern Lebanese U samples, we found only two individuals who carried haplotypes belonging to U5b2 and neither of these belonged to the U5b2c1 subgroup. It has been demonstrated, however, that modern Levantine populations do not reliably represent the haplogroups present in the Neolithic period, though haplogroup U5 was only found at low frequencies in pre-pottery Neolithic (PPNB) samples from the Levant [54]. U5b is found in only 0.4% of the modern populations from the Iberian peninsula, and only 0.18% Europe-wide [42], which provides us with further confidence of the authenticity of our ancient DNA result. Only the Saami, in northern Scandinavia, retain high levels of U5, and U5b1b in particular, with frequencies over 50% in some Saami populations [56]. Interestingly, haplogroup U5 and U5b have been identified in modern populations from North and Northwest Africa [57–59]. It must be noted, however, that there are few North African complete mitogenomes publicly available and HVR sequencing alone cannot identify beyond the U5b haplogroup (with the C150T HVR mutation) since the seven derived defining mutations (C1721T and A13637G, which define U5b2; A723G, 960XC and A13017G, which define U5b2c; and C6920a and A13484G, which define U5b2c1) are all in the coding regions. Achilli, et al. [59], using full mitochondrial genome sequencing identified a U5b1b1 cluster that grouped Amazigh (North African Berbers) and Saami populations. This cluster is based on the control region motif (16270–150) which is present at low frequencies in Amazigh, North African and nearly all European populations with the exception of the Scandinavian Saami where it is at about 48%. The divergence time of this cluster is around 8600 years ago (+/- 2400) consistent with an expansion from Franco-Cantabrian refuge which is believed to have been a major refuge for the European hunter-gatherers prior to their post LGM expansion [44]. It is suggested that European haplogroup U5 and the more prevalent U6 “Berber cluster” diverged from a common ancestor in the Near East and spread along the north and south coasts, respectively, of the Mediterranean, as far as Iberia to the north and Cyrenaica to the south [60]. It is very plausible that descendants of the Mesolithic hunter-gatherers carried U5b1b1 and sister lineages across the Straits of Gibraltar into North Africa [59], but there is no indication of when this migration may have happened. While it is possible that U5b2 haplogroups were also carried across the Straits of Gibraltar prior to Phoenician arrival in North Africa, our result now provides a minimum date of arrival. We can now say that U5b2c1 was present by the late 6th century BCE. So how did a young Phoenician man, with a European mitochondrial lineage end up in Carthage, North Africa? The earliest Phoenician site in Iberia is believed to be Gadir or Cadiz as it is known today, established in 1110 BCE [1]. Phoenician colonies were also established in Ibiza, southern Sardinia, western Sicily and along the southern shores of the Iberian Peninsula [1, 61, 62] and, later, these were also major Carthagenian trade ports. It has been suggested that the Phoenician east-west trade route from Tyre and Sidon across the Mediterranean and through the Straits of Gibraltar departed following a northerly route, travelling with the prevailing winds and tides westwards, stopping in Cyprus, Sicily, Ibiza and several locations along the southern coast of Spain, reaching Cadiz and beyond. Return trips back to the Levant took a southerly Mediterranean route, again, with the prevailing currents, sailing along the North African coast [1]. While early Phoenician occupation of and trade in the western Mediterranean would have been from the Levant, Carthage, once established, came to dominate the western Mediterranean trade networks, but would most likely have continued with this well travelled, counterclockwise circular route. Matisoo-Smith EA, Gosling AL, Boocock J, Kardailsky O, Kurumilian Y, Roudesli-Chebbi S, et al. (2016) A European Mitochondrial Haplotype Identified in Ancient Phoenician Remains from Carthage, North Africa. |
|
|
|
Post by Admin on Jun 16, 2016 22:40:17 GMT
The Mycenaean culture in ancient Greece commenced circa 1,650 BCE as an imported steppe culture from the northern Russian forest-steppes, which was known for the great mobility of its nomadic warriors. It is likely that the Mycenaeans originally migrated from South Russia to Greece between 1,900 and 1,650 BCE, where they intermingled with the locals, and their Y-DNA haplogroups were R1a, E-V13, G2a, I2a and J2. A following study on mitochondrial DNA from Grave Circle B in Mycenae (Hiller 1991) found that the ancient Mycenaean samples belonged to U5a1 or U5a1a and U5a lineages have also been found in Mesolithic Russia (U5a1) and Sweden (U5a1 and U5a2).  Type B, which has an oval shaft hole; six of them were found at Scetkovo, two at Kozorezovo (Pl. LV, c) and one at Berezan and Jekaterinoslav respectively 22. As to these specimens c.F. C. Hawkes in his still valuable article published in 1936 remarked that "in South Russia double axes appear in Bronze Age hoards directly recalling 'Treasure P' from the sixth city of Troy" 23. From Troy itself four double axes and one mould of corresponding shape are reported, yet only two of which have been published (Pl. LV, d-e) 24. Troy obviously took an active pan in the production of and the commerce with these tools. The same may be concluded from a celt mould which comes from Troy VII (Pl. LV, f) 25. It is of sorne interest here since celts were very popular in the North Pontic area as well as in the Balkan region whereas in Greece only few examples of that type of tool have been discovered 26.  Another group of implements which is found in Greece and the Northern Pontic area is represented by harnessing accessories. Here two pairs of bone cheek pieces from the Shaft Graves at Mycenae are paralleled by morphologically and structurally identical pieces from Trachtemirov near Kiev (Pl. LVI, a); this was recognized first by A. Leskov who also identified them as parts of horse-bits 27. The criticism raised against Leskov's interpretation by J. Crouwel and M.A. Littauer 28 has been convincingly refuted by H.-G. Hüttel who also discussed the problem of the relation between the Greek and the Ukrainian examples 29. Since, as he thinks, the chariot as a mean of warfare was not really essential for the steppe tribes to whom horse-riding was more adequate, the knowledge of this type of equipment reached the Ukraine from somewhere else; although a Near Eastern source is possible the close realationship between the Mycenaean and the Trachtemirov bits strongly recommends Mycenae as mediator 30.  This view is supported by a bone disc from Iljitevka (Krasnolimansk, Donec area) the ornament of which is closely related to that of a Mycenaean gold disc (Pl. LVI, b) 31. The higher quality of the Mycenaean piece points again to Mycenaean Greece as supplying the model. In this connection the spiral ornament on the pin-head from the Borodino Treasure 32 (found near Odessa) may be mentioned: it can be compared to similar designs on brooches from Mycenae (Pl. LVI, c); yet descent from a broader Balkan background remains no less possible; apart from that, no unanimity has been achieved so far among the experts as to the date of the hoard 33. That South Russia was reached by influences from the Balkan area carrying 'Mycenaean' elements with them is demonstrated by a "Stangen-Knebel" from Belz (Pl. LVI, d) near Sokal (Ukraine) 34 which exhibits the wide-spread circle and loop ornament, the main concentration of which is in Transylvania and Hungary; nevertheless it should be regarded as being originally Mycenaean 35. Regarding its wider distribution we may speak of a "Myceno- Balkanian koine" 36. For comparison a similar bone object from the IVth shaft grave at Mycenae may be referred to, the concrete function of which is however enigmatic 37  Into a broader horizon of cultural contacts, too, belongs the appearance of faience beads in South Russia as weIl as in the Kaukasus area; J. Bouzek has called it a "marginal area of distribution" 38, leaving open, however, 'marginal' to which centre; a Near Eastern one should, at least in my opinion, not be excluded 39, whereas J. Sulimirski, who refers to 157 faience beads found at Troy VI ("in the layer of ca. 1425 RC.") thinks that "faience beads were one of the commodities exported by the Trojans to Central Europe". And: "They attest well to the rôle of a Mycenaean agent played by the city and of the distributor of beads and other Mycenaean goods in the countries accessible by the Pontic route" 40. The question of the faience beads is, however, too vexed a problem as to be dealt within this connection.  Several authors felt that some steppe influence can be detected in the Early Mycenaean art, e.g. in the well known gold sheets from the 5th Shaft Grave showing wild animal in repoussée technique 54. This reminds us of the suggestion that also the boar tusks helmet is supposed to have originated in South Russia as it may be concluded from corresponding finds in several graves at Mariupol 55. Likewise the so-called arrow smootheners which appear in Greece for the first time during the later middle Helladic period have been referred to parallels in the Pontic area 56. This is also the case for horse burials; as S. Foltiny has pointed out, a characteristic feature of the cultures in the Volga-Ural area is the double burial of horses which is also known form Mycenaean Greece 57.  Shaft Grave showing wild animal is in repoussée technique 54. This reminds us of the suggestion that also the boar tusks helmet is supposed to have originated in South Russia as it may be concluded from corresponding finds in several graves at Mariupol 55. Likewise the so-called arrow smootheners which appear in Greece for the first time during the later middle Helladic period have been referred to parallels in the Pontic area 56. This is also the case for horse burials; as S. Foltiny has pointed out, a characteristic feature of the cultures in the Volga- Ural area is the double burial of horses which is also known form Mycenaean Greece 57. ln this relation special attention should be paid to an unusual kind of spiral ornament which is common to both a spear head from Cjurupinsk near Herson (Ukraine) and several shaft grave implements (Pl. LVII, c) 58. This ornament which is a newcomer to the Shaft Grave period has no immediate Aegean descent; could it have come from the region under discussion? I think there is no need to go as far as 1. Muhly who argued in favour of an immigration of steppe people to Greece at the beginning of the Shaft Grave period 59. But after all, it might be legal to wonder, as also J. Mulhy does 60, whether the gold from the Shaft Graves does not have really something to do with early Mycenaean contacts to the Pontic area which is well-known to be rich in gold; this is to be kept in consideration when a possible clue is looked for as to the legendary Golden Fleece 61. Hiller, Stefan. "The Mycenaeans and the Black Sea." Thalassa: L’Égèe prèhistorique et la mer: Actes de la troisième Recontre égéenne internationale de l’Université de Liège, Station de recherches sous-marines et océanographiques [StaReSo], Clavi, Corse, 23-25 avril 1990 (1991): 208-222. |
|
|
|
Post by Admin on Jul 3, 2016 22:40:01 GMT
Recently, mitochondrial DNA (mtDNA) from skeletal remains of European early farmers and late hunter-gatherers has been retrieved [7]–[13]. The frequency of mtDNA haplogroups, defined by substitutions shared by related mtDNA types (Phylotree.org-mtDNA tree build 12), in early farmers across Europe [7], [10]–[13] was found to be overall similar to those in modern Europeans (Figure 1, Figure S4, Figure S5), while pre-Neolithic hunter-gatherers appear to be quite distinct (Figure 1). In particular, 83% (19 out of 23) of hunter-gatherers analyzed to date carry mtDNAs belonging to haplogroup U [9], [10], [14] and none of the hunter-gatherers fall in haplogroup H. In contrast, haplogroup U has been found in only 13 of 105 (around 12%) individuals from early farming cultures of Europe and it occurs in less than 21% of modern Europeans, while haplogroup H comprises between 25% and 37% of mtDNAs retrieved from early farming cultures (Figure S4) and is in about 30% of contemporary Europeans (Figure 1). The mtDNA data thus suggest that the pre-Neolithic populations in Europe were largely replaced by in-coming Neolithic farming groups, with a maximum mtDNA contribution of around 20% from pre-Neolithic hunter-gatherers [8]–[10]. The genetic contribution of pre-Neolithic hunter-gatherers to later Neolithic populations is furthermore supported by a similar frequency of U subhaplogroups (U5, U4, K and U2) that were found in pre-Neolithic hunter-gatherers (Figure S3) and are still the most common U-subhaplogroups in modern Central Europeans (Figure S5).  Whereas H-type mtDNAs have on average six nucleotide differences in their coding region (position 577–16023) (Figure 2, green), U-type mtDNAs have on average 18 differences (Figure 2, red). The distribution of pair-wise differences among the H-type mtDNAs shows a clear mode around 6 differences whereas the U-types have a mode around 22 differences. Such peaks may be caused by past population expansions [20] (Figure S7, Figure S8, Figure S9). They would suggest that H-type mtDNAs experienced a recent population expansion while U-type mtDNAs experienced a much older population expansion. Notably, these differences in the distributions of pair-wise nucleotide differences are not caused by sequencing of a selected set of mtDNA types present in GenBank, since dataset 2 as well as the individuals sequenced from Croatia (dataset 3) show an average number of differences as well as modes very similar to dataset 1.  In order to analyze potential population size changes over time, we calculated Bayesian skyline plots using the BEAST package [21] for dataset 1 and dataset 2 (dataset 3 was too small). In both datasets, the direct comparison of skyline plots between the H-type and the U-type mtDNAs (Figure 3) reveals a population increase for individuals carrying the H-type starting around 9,000 YBP and continuing to the present, whereas the U-type shows a population expansion between 20,000 and 10,000 YBP with a putative period of slight decrease between 6,000 and 5,000 YBP (Figure S6A, B). For both U-type and H-type mtDNAs, we observe similar patterns of population growth starting around 4,000 YBP to the present (Figure 3). Thus, H-type and U-type mtDNAs show a distinct population history before 5,000 YBP, possibly reflecting that they were primarily present in different populations with different origins and histories.  The high frequency of H-type mtDNAs in European Neolithic populations and its complete absence in pre-Neolithic hunter-gatherers suggests that H-type mtDNAs arrived with early farmers in Europe. The population size increase observed between 9,000 and 5,000 YBP likely represents the population expansion that accompanied the Neolithic revolution. In contrast, U-type mtDNAs show an increase in population size around 15,000 to 10,000 YBP, which coincides with the end of the last glacial maximum in Europe and a northwards expansion of hunter-gatherer populations. The data suggests that this population remained rather constant after 10,000 YBP until the onset of the Neolithic revolution. However, the H-type mtDNA population size seems to experience an exponential increase around 7,000 YBP, suggesting that both populations are not yet fused. After 4,000 YBP, no archaeological remains of hunter-gatherers were found in central Europe [22]. From approximately that time on, both H- and U-type mtDNAs expand in a similar way. This may reflect fusion of the two populations where these mtDNAs were prevalent.  These results suggest that H-type mtDNAs in the European mtDNA gene pool show evidence of a population expansion related to the spread of animal husbandry and farming. In contrast, U-type mtDNAs seem to represent earlier hunter-gatherers that adopted farming practices and admixed with immigrant farming populations. In agreement with this scenario, the only non-agricultural population of Europe, the Saami in Northern Scandinavia and Russia, carry about 49% of U-type mtDNAs [23]. Fu Q, Rudan P, Pääbo S, Krause J (2012) Complete Mitochondrial Genomes Reveal Neolithic Expansion into Europe. PLoS ONE 7(3): e32473. doi:10.1371/journal.pone.0032473 |
|













