|
|
Post by Admin on Jan 13, 2018 18:55:49 GMT
 The population of the UK today is culturally diverse, with 8% of its 54 million inhabitants belonging to ethnic minorities, and over 1 million classifying themselves as ‘Black or Black British’ in the 2001 census. These people owe their origins to immigration from the Caribbean and Africa beginning in the mid twentieth century; prior to this time, the population has been seen as typically Western European, and its history has been interpreted in terms of more local immigration, including that of the Saxons, Vikings and Normans1. However, in reality Britain has a long history of contact with Africa (reviewed in ref. 2). Africans were first recorded in the north 1800 years ago, as Roman soldiers defending Hadrian's wall – ‘a division of Moors’. Some historians suggest that Vikings brought captured North Africans to Britain in the 9th century. After a hiatus of several hundred years, the influence of the Atlantic slave trade began to be felt, with the first group of west Africans being brought to Britain in 1555. African domestic servants, musicians, entertainers and slaves then became common in the Tudor period, prompting an unsuccessful attempt by Elizabeth I to expel them in 1601. By the last third of the eighteenth century there were an estimated 10,000 black people in Britain3, mostly concentrated in cities such as London. Has this presence left a genetic trace among people regarded as ‘indigenous’ British? In principle, Y-chromosomal haplotyping offers a means to detect long-established African lineages. Haplotypes of the non-recombining region of the Y, defined by slowly mutating binary markers such as SNPs, can be arranged into a unique phylogeny4-6. These binary haplotypes, known as haplogroups (hg), show a high degree of geographical differentiation, reflecting the powerful influence of genetic drift on this chromosome. Some clades of the phylogeny are so specific to particular continents or regions that they have been used to assign population-of-origin to individual DNA samples7, and in quantifying the origins of the components of admixed populations using simple allele-counting methods8-10.  Studies of British genetic diversity, generally sampling on the criterion of two generations of residence, have found no evidence of African Y-chromosomal lineages11-14, suggesting either that they never became assimilated into the general population, of have been lost by drift. However, here we describe a globally rare and archetypically African sub-lineage in Britain and show that it has been resident there for at least 250 years, representing the first genetic trace of an appreciable African presence that has existed for several centuries2. As part of a survey of British Y chromosome diversity, we recruited a set of 421 males who described themselves as British, and whose paternal grandfathers were born in Britain. The Y chromosomes of these males were typed using a set of 11 binary markers15, including M145 (defining superhaplogroup DE), and M89 (defining superhaplogroup F). All chromosomes carried the derived allele at one or other of these two markers, with a single exception, in male GB1757, which could in principle belong to hgA, B, or C (see phylogeny in Figure 1). Further testing, including the markers M91 and M31, gave the surprising result that it belonged to hgA, within the sub-lineage A1. 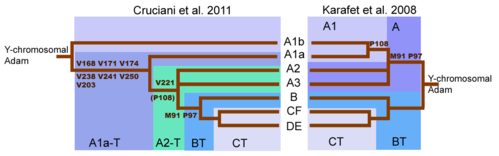 |
|
|
|
Post by Admin on Jan 15, 2018 19:08:50 GMT
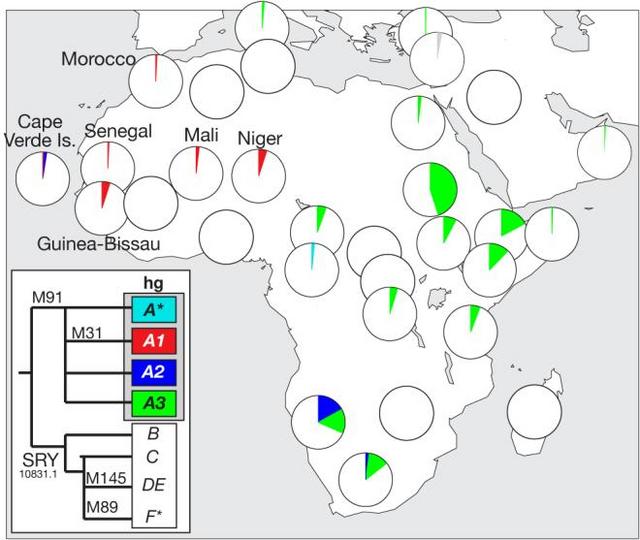 Figure 1 Distribution of Y chromosomes belonging to haplogroup A Haplogroup A is the deepest-rooting clade of the Y phylogeny, and shows a particularly specific localization to the African continent (Figure 1), which is compatible with an African origin for modern human Y chromosomes. It constitutes 5.4% of a composite sample of 3551 Africans4,8,10,24-30, while in non-African indigenous populations only seven cases have been described, from Turkey31, Cyprus32, Sardinia33,34 and Oman35. Extensive surveys of western European populations have failed to find any examples of these chromosomes13,36-39. The sub-haplogroup A1 was first reported in a single individual among a sample of 44 males from Mali4. Subsequently, this scarce western African haplogroup has been found in only 25 more males (Figure 1): 2/64 Moroccan Berbers24, 3/766 African-Americans40,41, 2/39 Mandinka from Gambia/Senegal, 1/55 Malian Dogon30, 1/201 Cape Verde Islanders, 14/276 males from Guinea-Bissau10, and 2/39 males from Niger (F.C. and R.S., unpublished data). The British male carrying the hgA1 chromosome knew of no familial African connection, and he displays a typical European appearance. To investigate the relationship of his Y chromosome with African examples, we compared its 10-locus Y-STR (short tandem repeat) haplotype (see Supplementary information) with those of ten other available hgA1 chromosomes10,24,40 (F.C. and R.S., unpublished data). Figure 2a shows a median-joining network of these haplotypes: they are diverse, and all are unique. Though the British haplotype is peripheral, it lies equidistant (4 mutational steps) from Niger and Guinea-Bissau haplotypes, and similar distances (2-4 steps) exist between other haplotypes in the network. This is compatible with a western African origin for the British chromosome, but does not point to a particular population. Using the British haplotype (11 loci) to search the Y Chromosome Haplotype Reference Database ( www.yhrd.org) finds no matches among 15,815 chromosomes worldwide, emphasising its rarity. Also, when the haplotypes of the other hgA1 chromosomes are used in similar searches, they find only self-matches in the populations from which they derive, underlining the scarcity and African-specificity of hgA1. 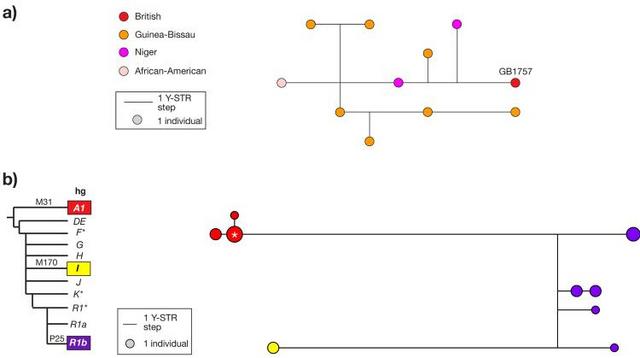 Figure 2 Diversity of Y-STR haplotypes of chromosomes belonging to haplogroup A1, and within the R surname How long has this archetypically African Y chromosome been in England? To address this question our strategy was to seek patrilinearly related individuals who would share the haplogroup, but whose Y-STR haplotype diversity could be used to estimate a time-to-most-recent-common-ancestor (TMRCA). To do this we exploited the relationship between surnames and Y chromosome haplotypes15,42-44, noting that the upper bound of any estimated age would be limited by the fact that hereditary English surnames did not exist prior to the eleventh century45. The hgA1-bearing male bears a locative surname, which we refer to here as R, deriving from an East Yorkshire village46. Only 121 people carried this name in 1998 ( www.spatial-literacy.org/uclnames), and it still has a strong east Yorkshire focus. We recruited 18 apparently unrelated men carrying this name (or a close variant spelling, carried by 50 individuals) and typed a set of 11 binary markers and 17 Y-STRs15, supplemented with the binary marker M31, allowing us to identify hgA1. Figure 2b shows a median-joining network of 17-locus Y-STR haplotypes (see Supplementary information) of Y chromosomes carried by 18 R-surnamed males. The chromosomes belong to three haplogroups, and include four clusters, indicating either multiple foundation or historical nonpaternity within the name. However, a total of seven of the males carry hgA1 chromosomes, belonging to three closely related Y-STR haplotypes, and, based on the rho statistic within Network, having a TMRCA of 440 ± 330 years. |
|
|
|
Post by Admin on Jan 16, 2018 19:09:10 GMT
As an empirical adjunct to TMRCA calculations, we undertook extensive genealogical research to ask if the seven R-surnamed males carrying hgA1 chromosomes could be connected into a single genealogy with a historically verifiable MRCA. This research resolved the males into two well-supported genealogies (Figure 3a), with MRCAs born in 1788 and 1789 respectively. However, although both of these ancestors were resident in Yorkshire, evidence could not be found for a familial relationship between them. Patterns of forename usage in the two genealogies are quite distinct, which argues against a very recent connection. We recruited 12 unrelated R-surnamed men from the USA, hoping that presence of hgA1 would indicate that the chromosome had been associated with the surname prior to emigration from Britain: however, none of these men carried a chromosome from this haplogroup (data not shown), so the approach was uninformative.  Figure 3 Genealogical relationships and Y-STR haplotypes of hgA1 R-surnamed men Discussion Our study shows that a globally rare Y chromosome type, belonging to the deepest-rooting African branch of the Y phylogeny, has been present in northern England since at least the mid eighteenth century. Haplogroup E3a is by far the most frequent Y-chromosomal lineage in Africa, existing at 48% in a continent-wide sample of 1122 chromosomes30, so we would expect any substantial past immigration from Africa to Britain to have left examples of chromosomes belonging to this common haplogroup. However, a survey of 1772 Y chromosomes from the British Isles found none13, and they are also absent from our control sample of 421 chromosomes. The general rarity of African lineages may reflect a low level of initial introgression, later loss through drift, or sampling bias – for example, the large British survey13 sampled from small towns, in which the descendants of early British Africans, who were concentrated in cities, may be depleted. Admixture between populations of African and European origin is often sex-biased, with a greater proportion of the African component of the hybrid population being contributed by females48. Assuming an equal number of males and females of African origin migrating to Britain, we might therefore expect mitochondrial DNA to reveal a stronger signal of African admixture than the Y chromosome. There is little published evidence, but a study of mitochondrial DNA sequence diversity among 100 ‘white Caucasian’ British49 does contain one haplotype which represents a haplogroup L1c sequence (defined according to ref. 50), with a probable origin in west Central Africa51. This could represent a possible maternal counterpart to the Y lineage we describe here. The presence of a very rare type of Y chromosome in two distinct branches of a genealogy that coalesce prior to the late eighteenth century demonstrates clearly that the chromosome must have been introduced before this time. However, the upper limit on the time of linkage with the R surname is less certain. In principle the association could have been formed many generations earlier, with the descendants of any earlier-branching lineages having drifted to extinction, or not sampled in our study. Since the pattern of Y diversity in the surname suggests either multiple origins for the name, or nonpaternities introducing diverse Y chromosomes, the hgA1 lineage need not have been a founding type at the time of surname establishment (about 700 years ago45). The contributor of the chromosome to the surname may himself have been a first-generation immigrant African, or an admixed and phenotypically European man carrying an African Y chromosome introduced into Britain some time earlier. The remarkable Y chromosome present in the R surname provides the first genetic evidence of a long-lived African presence within Britain. It represents a cautionary tale for those who would predict population-of-origin from a Y haplogroup7, and emphasises the complex nature of the history of human migration. Eur J Hum Genet. 2007 Mar; 15(3): 288–293. Published online 2007 Jan 24. doi: 10.1038/sj.ejhg.5201771 |
|
|
|
Post by Admin on Feb 3, 2018 18:46:58 GMT
 Researchers from the Grantham Centre for Sustainable Futures at the University of Sheffield have shed light on how hunter-gatherers first began farming and how crops were domesticated to depend on humans. Domesticated crops have been transformed almost beyond recognition in comparison with their wild relatives -- a change that happened during the early stages of farming in the Stone Age. For grain crops like cereals, the hallmark of domestication is the loss of natural seed dispersal -- seeds no longer fall off plants but have become dependent on humans or machines to spread them. Professor Colin Osborne, from the Grantham Centre for Sustainable Futures at the University of Sheffield, said: "We know very little about how agriculture began, because it happened 10,000 years ago -- that's why a number of mysteries are unresolved. For example why hunter-gatherers first began farming, and how were crops domesticated to depend on people. "One controversy in this area is about the extent to which ancient peoples knew they were domesticating crops. Did they know they were breeding domestication characteristics into crops, or did these characteristics just evolve as the first farmers sowed wild plants into cultivated soil, and tended and harvested them?" The new research, published in the journal Evolution Letters, shows the impact of domestication on vegetable seed size. Any selective breeding of vegetables by early farmers would have acted on the leaves, stems or roots that were eaten as food, but should not have directly affected seed size. |
|
|
|
Post by Admin on Apr 4, 2018 18:50:05 GMT
Jewish surnames have not been different in their evolution and, among others, are currently comprised of Aramaic and Hebrew patronymic surnames, surnames representing a Diaspora residency or occupation, and modern day Hebraized names11,12. World Jewry, estimated at 14 million individuals, can be roughly divided into Ashkenazi and non-Ashkenazi Jews. The former group is considered to have been formed approximately 2,000 ybp and to account for approximately 75% of contemporary Jews13. Notably, despite this remote split, Ashkenazi and non-Ashkenazi communities share two designations representing the two Jewish priesthood lineages, Levite (Levi, in Hebrew) and Cohen, whose etymologies relate to the Biblical male ancestors Levi and Aaron14. According to the Biblical narrative, Levi was the third son of the Biblical Patriarch Jacob. Levi’s given name was transformed, in time, to the Levite tribal honorific Halevi (The Levite, in Hebrew). One great-grand son of Levi, namely Aaron, was given an honorific to represent his occupation – Cohen (to serve, in Hebrew), the first high priest. Alternative theories for the origins of the Levite caste have been proposed15. While many contemporary Levites still use the Biblical surname form (Levi), the surname continued to evolve throughout the millennia through phonetic spelling variations (e.g. Levin, Lewicki), or through the adoption of a residency location of a specific Levite dynasty as a surname. An illustrative example of the latter is the Horowitz Rabbinical Levite dynasty16 established by the migration of one Levite family from Girona, Catalonia to Horovice, a small town near Prague, Czech Republic, circa 1400 CE. While claims for documented origin in medieval Spain have been made, the founder of the dynasty is considered to be Yeshayah ‘Horovsky’ Ish Horovice (1450–1514 CE)17. Genealogical records of the Horowitz patrilineal dynasty comprising no less than 15 subsequent generations are available17.  Not surprisingly, the Levite and Cohen castes have been the focus of a series of genetic studies during the past two decades using ever-expanding portions of the Y chromosome for the analysis18,19,20,21,22. First, the Cohen dynasty was studied and found to have a limited number of founding lineages that were shared between Ashkenazi and non-Ashkenazi Jews19,21,22. The most frequent Cohen lineage, comprising 46.1% of contemporary, self-identifying Cohen males, is found within haplogroup J1-P58, which is prevalent in the Middle East19. Next, it was shown that the paternal ancestry found among Ashkenazi Levites is dominated by a set of tightly evolving Y chromosome lineages falling within haplogroup R1a-M198 which was, at the time of publication, the most resolved branch known on this evolutionary path18. Other haplogroups reported among Ashkenazi Levites demonstrated no additional significant founding event, and the haplogroup R1a-M198 founder event was not shared with Sephardi Levites. These findings captured the attention of both scientists and laypersons, as the magnitude of the founder effect suggests that fully ~200,000 males with the tradition of Levite descent share a recent common direct male ancestor within recent historical time frames23. Importantly, the initial genetic analyses suggested in this first publication incorrectly attributed this Ashkenazi Levite lineage’s origin to Eastern Europe18. A follow up study, summarizing information from whole Y chromosome sequencing, focused specifically on this Ashkenazi Levite lineage and confirmed that that 65% of the 97 randomly assembled Ashkenazi Levites carried haplogroup R1a-M19820. Strikingly, the better resolved whole Y chromosome based phylogeny of haplogroup R1a, showed that 100% of these samples could be reassigned to the refined haplogroup R1a-M582. This distinctive R1a-M582 lineage was found, other than in Ashkenazi Jews, among 15.7% males self-affiliating as non-Ashkenazi Levites and, importantly, at low frequencies only in the Middle East, consistent with this location as its ancestral origin20.  While the phylogenetic origin of the R1a-M582 lineage was clarified20, the aim of this study is to further explore several questions that remained open regarding this founder lineage among Ashkenazi Levites. First, the limited number of whole Y chromosome sequences from Ashkenazi Levites had precluded a definitive description of their phylogenetic branch, its coalescence time and its route of entrance to Europe (Fig. 1). Second, the ancestral ties between Ashkenazi Jews self-affiliating as Levites and Ashkenazi Jews self-affiliating as non-Levites within haplogroup R1a-M582 remained elusive. Third, the existence of haplogroup R1a-M582 in both Ashkenazi and non-Ashkenazi Levites was not explained. Fourth, lack of whole Y chromosome sequences from other Ashkenazi haplogroups did not allow a comparison between a specific founding event for Ashkenazi Levites and a general expansion of Ashkenazi Jews that also affected the Ashkenazi Levites. Fifth, a comparison between the coalescence ages of the dominant Cohen priesthood J1-P58 lineage shared between Ashkenazi and non-Ashkenazi Jews and the Ashkenazi Levite lineage at the level of the whole Y chromosome sequences was yet to be conducted. Having these objectives in mind, we assembled 486 whole Y chromosome sequences from Ashkenazi Jews with a tradition of Levite descent (Supplemental Table S1), including members of the Horowitz rabbinical dynasty, Ashkenazi Jews without a tradition of Levite descent, non-Ashkenazi Jews and non-Jews. Of these, 179 are novel, including 65 R1a-M582 samples that were collected following expert genealogical input. This set of 65 samples consists of males with 56 different surnames, who claim to have an Ashkenazi Levite paternal origin. Samples were chosen to include the widest possible range of haplogroup R1a-M582 internal variation based on their previously available short tandem repeat (STR) haplotypes (Supplemental Table S2). Additional samples were included to provide the appropriate phylogenetic framework for the studied haplogroups. |
|






