|
|
Post by Admin on Dec 4, 2013 1:15:59 GMT
 Many authors have speculated on Nubian biological evolution. Because of the contact Nubians had with other peoples, migration and/or invasion (biological diffusion) were originally thought to be the biological mechanism for skeletal changes in Nubians. Later, a new hypothesis was put forth, the in situ hypothesis. The new hypothesis postulated that Nubians evolved in situ, without much genetic influence from foreign populations. This study examined 12 Egyptian and Nubian groups in an effort to explore the relationship between the two populations and to test the in situ hypothesis. Data from nine cranial nonmetric traits were assessed for an estimate of biological distance, using Mahalanobis D(2) with a tetrachoric matrix. The distance scores were then input into principal coordinates analysis (PCO) to depict the relationships between the two populations. PCO detected 60% of the variation in the first two principal coordinates. A plot of the distance scores revealed only one cluster; the Nubian and Egyptian groups clustered together. The grouping of the Nubians and Egyptians indicates there may have been some sort of gene flow between these groups of Nubians and Egyptians. However, common adaptation to similar environments may also be responsible for this pattern. Although the predominant results in this study appear to support the biological diffusion hypothesis, the in situ hypothesis was not completely negated. The Mahalanobis D2 analysis uncovered close affinities between Nubians and Egyptians. Table 3 lists the Mahalanobis D2 distance matrix. As there is no significance testing that is available to be applied to this form of Mahalanobis distances, the biodistance scores must be interpreted in relation to one another, rather than on a general scale. In some cases, the statistics reveal that the Egyptian samples were more similar to Nubian samples than to other Egyptian samples (e.g. Gizeh and Hesa/Biga) and vice versa (e.g. Badari and Kerma, Naqada and Christian). These relationships are further depicted in the PCO plot (Fig. 2). Aside from these interpopulation relationships, some Nubian groups are still more similar to other Nubians and some Egyptians are more similar to other Egyptian samples. Moreover, although the Nubian and Egyptian samples formed one well-distributed group, the Egyptian samples clustered in the upper left region, while the Nubians concentrated in the lower right of the plot. One line can be drawn that would separate the closely dispersed Egyptians and Nubians. The predynastic Egyptian samples clustered together (Badari and Naqada), while Gizeh most closely groups with the Lisht sample. The first two principal coordinates from PCO account for 60% of the variation in the samples. The graph from PCO is basically a pictorial representation of the distance matrix and interpretations from the plot mirror the Mahalanobis D2 matrix. Discussion  PCO plot of biological affinities. PCO plot of biological affinities.The clustering of the Nubian and Egyptian samples together supports this paper’s hypothesis and demonstrates that there may be a close relationship between the two populations. This relationship is consistent with Berry and Berry (1972), among others, who noted a similarity between Nubians and Egyptians. If Nubians and Egyptians were not biologically similar, one would expect the scores to separately cluster by population (e.g. Nubians compared to Nubians would have small biological distances, and Nubians compared to Egyptians would have high biological distances). However, this was not the case in the current analysis and the results suggest homogeneity between the two populations. Many of the samples that are similar to one another, between the two populations, are separated by great amounts of time (e.g. Kerma and Badari). These similarities over time make sense because, as Konigsberg (1990) asserted, as time elapses, related groups become more genetically similar. In order to explicate the meaning behind all of these findings, the results here must be tempered by the DNA evidence. Both mtDNA (Krings et al., 1999) and Y-Chromosome data (Hassan et al., 2008; Keita, 2005; Lucotte and Mercier, 2003) indicate that migrations, usually bidirectional, occurred along the Nile. Thus, the osteological material used in this analysis also supports the DNA evidence. Interpretation of the results framed by several of the groups’ histories helps to elucidate the subtle relationships depicted in the PCO scatter plot. The predynastic sample from Badari occupies a complex position in Egyptian history. The Badarians are Egypt’s oldest agriculturalists and produced some of the earliest known pottery (Hassan, 1986) that predated state formation in Egypt. Badarian crania, in comparison to dynastic groups, are slight and less robust than their later counterparts (Angel, 1972; Morant, 1935; Stoessiger, 1927). Stoessiger (1927) likened the gracile nature of the Badarians to the gracile nature of the people from Naqada, but she pointed out that the Badarians are more prognathic. On this basis, many have postulated that the Badarians are relatives to South African populations (Morant, 1935 G. Morant, A study of predynastic Egyptian skulls from Badari based on measurements taken by Miss BN Stoessiger and Professor DE Derry, Biometrika 27 (1935), pp. 293–309.Morant, 1935; Mukherjee et al., 1955; Irish and Konigsberg, 2007). The archaeological evidence points to this relationship as well. (Hassan, 1986) and (Hassan, 1988) noted similarities between Badarian pottery and the Neolithic Khartoum type, indicating an archaeological affinity among Badarians and Africans from more southern regions. Furthermore, like the Badarians, Naqada has also been classified with other African groups, namely the Teita (Crichton, 1996; Keita, 1990), while the Gizeh sample clustered with the Maghreb and Sedment (Dynasty IX Egyptians) (Keita, 1990). Nutter (1958) noted affinities between the Badarian and Naqada samples, a feature that Strouhal (1971) attributed to their skulls possessing “Negroid” traits. Keita (1992), using craniometrics, discovered that the Badarian series is distinctly different from the later Egyptian series, a conclusion that is mostly confirmed here. In the current analysis, the Badari sample more closely clusters with the Naqada sample and the Kerma sample. However, it also groups with the later pooled sample from Dynasties XVIII–XXV. The unusual grouping of Badari, Naqada, Kerma, and the later Dynastic pooled sample may have been a product of the mixed nature of the pooled sample. The effects of pooled samples have been demonstrated in Nubians by obscuring relationships and creating a falsely close affinity between it and the samples it clusters with (Godde, 2009a). Moreover, affinities among the Badarian, Naqada, and Kerma samples have been revealed by other authors (Keita, 1990; Nutter, 1958) and it is no surprise that this relationship exists in the data here. Relationships among Badari, Naqada, and Kerma have not always been overt in the skeletal data. Berry et al. (1967) concluded from their nonmetric analysis that their Badarian sample differed significantly from Naqada and Kerma, but was closely related to the Gizeh sample. Their study included the same samples as this analysis, but yielded results that are different from the current study and the craniometric research. Berry et al. (1967) employed a completely different range of statistics, which may account for the difference between the two conclusions. However, Berry and her coauthors also noted homogeneity across all the Egyptian groups, including Naqada and those that pre- and post-date the sample. This is indeed the case here, as is evidenced in the PCO plot; the Egyptians appear to be relatively homogeneously grouped. Some Badarian crania also classified well with the Gizeh sample (Keita, 1990). The close clustering of Badari and Naqada with Kerma exemplifies the possible relationship of Nubians to Egyptians. Originally, the Nubian A-Group was thought to be Badarian in origin (Reisner, 1910). However, later work (Adams, 1977; Godde, 2009a) established that the A-Group were actually Nubian. Comparisons of C-Group and Pan-Grave Nubians to Badari and Hierakonpolis separate Badari from the other samples, indicating no biological affinities with these earlier Nubian groups (Godde, 2009b). The reoccurring notation of Kerma affinities with Egyptian groups is not entirely surprising. Kerma was an integral part of the trade between Egypt and Nubia. Collett (1933) concluded that Kerma was originally inhabited by Egyptians with neighboring Nubian settlements. Her investigation of the site pointed towards continuous Egyptian occupation of some sort at the site throughout the Kerma time period. This continued presence at Kerma is an optimal condition for gene flow to occur between the two populations. Nubian groups have also been scrutinized as to their relationship with other Nubians. Both the Meroitic and X-Group were originally postulated to be foreign peoples migrating into Lower Nubia (Adams, 1968; Nielsen, 1970). These ideas were based on changes in pottery around the beginning of each of the respective time periods. However, the archaeological evidence actually showed slow change in form over time (Adams, 1977) and the biological evidence demonstrated a similar trend in the skeletal data (e.g. Godde, in press; Van Gerven et al., 1977). These conclusions negate the possibility of invasion or migration causing the shifts in time periods. The results in this study are consistent with prior work; the Meroites and X-Group cluster with the remaining Nubian population and are not differentiated.  Despite the biological similarities between the two populations, the Nubians appear relatively homogeneous. The homogeneity is consistent with Carlson and Van Gerven’s (1979) in situ hypothesis, but contradicts the findings of Buzon (2006). Buzon (2006) found a high level of heterogeneity in the Nubian samples she examined, including individuals from Kerma and the C-Group. Moreover, the Egyptian samples in her study were homogeneous overall, consistent with Berry et al. (1967) and the results in this paper. However, the levels of homogeneity appear to be similar within Nubians and within Egyptians in this study. The differences between this research and Buzon’s (2006) work may be related to the statistics used. Buzon’s (2006) goal was not to look at biological affinities; rather, she was trying to establish identity among her individuals by associating it with archaeological material. While this paper used a biological distance approach to investigate past population relationships, her paper used factor analysis, principal components, and a least squares regression. Although these (hers and those used here) statistics all have a solid methodological basis, they measure population relationships in two different manners and the results between them are not entirely comparable. Gene flow may account for the homogeneity across these Nubian and Egyptian groups and is consistent with the biological diffusion precept. Small geographic distances between groups allow for the exchange of genes. One of the Nubian groups in this analysis is located in Upper Egypt (Hesa/Biga), near Egyptian occupation, and contact between the two populations may have been commonplace. Specifically, Nubians were often captured and enslaved by Egyptians to build pyramids, or employed by the Egyptian army (Trigger, 1976). Occasionally, Nubians were even directed to fight other Nubians as part of their duties as troops (Trigger, 1976). Moreover, some groups of Nubians allied with the Egyptians for the conquest of Nubian areas, primarily during Dynasty I (Trigger, 1976). Furthermore, as mentioned earlier, trade between Nubians and Egyptians flourished at Kerma and Meroe, during the time periods named after the sites, and enabled contact for potential gene flow. As a result of their respective histories, the multitude of interactions between them, geographic locations, and their biological composition, it appears that gene flow was possibly occurring between the two populations. The similarities uncovered by this study may be explained by another force, adaptation. As stated above, the results appear to support the biological diffusion hypothesis because the Nubian and Egyptian groups are biologically similar. However, this resemblance may be indicative of a common adaptation to a similar geographic location, rather than gene flow. Carlson and Van Gerven (1979) stated this idea in reference to common adaptations of Nubian, Paleolithic, and aboriginal Australian populations. Additionally, Carlson (1976), Prowse and Lovell (1995), Van Gerven (1982), and Van Gerven et al., 1977 D. Van Gerven, G. Armelagos and A. Rohr, Continuity and change in cranial morphology of three Nubian archaeological populations, Man 2 (1977), pp. 270–277. View Record in Scopus | Cited By in Scopus (9)Van Gerven et al. (1977) also recognized this form of natural selection as a mechanism for in situ biological change; Egypt and Nubia have similar terrain and climate. Because of the similarity between and the overlapping of the two territories that would require similar adaptations to the environment, common adaptation cannot be discounted. In summation, a portion of the in situ hypothesis in Nubians is supported in this paper, namely homogeneity. Gene flow appears likely between the Egyptians and Nubians, although common adaptations to a similar environment may have also been a factor in their cranial similarities. This study does not rule out the possibility that in situ biological evolution occurred at other times not represented by the samples in this analysis. Further research should incorporate more populations the Nubians were in contact with, to further shed light on Nubian population structure. Additionally, Konigsberg’s (1990) spatial–temporal isolation model should be applied to the dataset here to further explicate the results. Godde, K. "An examination of Nubian and Egyptian biological distances: Support for biological diffusion or in situ development?." HOMO-Journal of Comparative Human Biology 60.5 (2009): 389-404. |
|
|
|
Post by Admin on Dec 5, 2013 15:52:26 GMT
 Excavations of a complex of caves in the Sierra de Atapuerca in northern Spain have unearthed hominin fossils that range in age from the early Pleistocene to the Holocene1. One of these sites, the ‘Sima de los Huesos’ (‘pit of bones’), has yielded the world’s largest assemblage of Middle Pleistocene hominin fossils2, 3, consisting of at least 28 individuals4 dated to over 300,000 years ago5. The skeletal remains share a number of morphological features with fossils classified as Homo heidelbergensis and also display distinct Neanderthal-derived traits6, 7, 8. Here we determine an almost complete mitochondrial genome sequence of a hominin from Sima de los Huesos and show that it is closely related to the lineage leading to mitochondrial genomes of Denisovans9, 10, an eastern Eurasian sister group to Neanderthals. Our results pave the way for DNA research on hominins from the Middle Pleistocene. DEEP inside the Atapuerca cave system in northern Spain, 30 metres beneath the surface, lies the Sima de los Huesos, or the "pit of bones". The remains of at least 28 ancient humans have been found at the bottom of this 12-metre-long vertical shaft. Now a thigh bone pulled out of the pit has yielded 400,000-year-old DNA – by far the oldest human DNA ever sequenced. The results suggest the thigh bone belonged to a previously unknown human species – perhaps even a missing link between the Neanderthals and their mysterious cousins the Denisovans. This, say palaeontologists, brings us closer than ever before to understanding who our own common ancestor with the Neanderthals was. The bones at Sima de los Huesos pre-date the origin of Homo sapiens, who appeared around 200,000 years ago, and most closely resemble those of Neanderthals. Fred Spoor of the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany, calls them "Neanderthals in the making". Until now, it had only been possible to sequence the genomes of hominin fossils found in cold climates; DNA breaks down faster in warmer climates like Spain's. But spurred by the successful sequencing of a 300,000-year-old cave bear genome from the same area, Matthias Meyer, also at the Max Planck Institute in Leipzig, and colleagues decided to give it a go. They drilled into a hominin thigh bone from the cave and extracted 1.95 grams of material, processed it for DNA, and filtered out a large amount of modern human DNA – the bones had been heavily contaminated as they were removed and handled. The end result was a near-complete mitochondrial genome – the DNA found inside the organelles that power cells. By comparing it with that of modern humans, chimpanzees and bonobos, plus Neanderthals and Denisovans, Meyer estimated its age at 400,000 years, twice as old as our own species and far older than any hominin genome previously sequenced ( Nature, DOI: 10.1038/nature12788). The Neanderthal and Denisovan genomes sequenced in recent years are each around 40,000 years old.  The skeleton of a hominin recovered from Sima de los Huesos. The skeleton of a hominin recovered from Sima de los Huesos.The biggest mystery is how and when our lineage diverged from that of the Neanderthals and Denisovans. Also unclear are the circumstances of the later split between Neanderthals and Denisovans. All we know is that both of these events happened around the time the Sima de los Huesos hominins were living in Spain. One possibility is that the fossils belong to the common ancestor of Neanderthals and Denisovans, and some of their descendants later headed east and became the Denisovans. The Sima de los Huesos genome is particularly exciting because it is from a time that is very close to the origin of our human line. The archaeological evidence suggests these early humans were developing significant new behaviours. On the one hand, they were still using fairly primitive stone tools like a crafted hand axe – nicknamed Excalibur – that was found in the pit. But the bones also suggest more modern traits.  A 2012 study on an early modern human from Tianyuan Cave, China ( Pääbo et al. 2012) detected the minute presence of Denisovan genetic material in European groups (0-0.2%) and Denisovans may have left their genetic footprint on both Europeans and Asians. A hominid discovered at the ‘Sima de los Huesos’ is a Denisovan individual with Neanderthal features and Neanderthals and Denisovans may have interbred in Europe before H. sapience arrived and Denisovan DNA was passed on to present-day Europeans and East Asians through Neanderthals without direct Denisovan/H. sapience admixture, while Denisova gene flow occurred into the common ancestors of Melanesian groups (1.7-4.7%). |
|
|
|
Post by Admin on Dec 13, 2013 5:25:28 GMT
Even if archaeological and paleoanthropological records testify to the ancient (Paleolithic) human occupation of North Africa, the evolution of human groups living in that area is still unclear. This is mainly due to the large number of successive prehistoric and historic events that occurred in that area after the arrival of the first modern humans. In North Africa, the presence of Berbers – a term to denote those populations which speak a Berber language (Camps 1980) – is well described since the Capsian (10,000–4700 years ago), although this industry derived from oldest cultures. Nevertheless, it was only during the Neolithic transition (around 6000 years ago in the Saharan areas and 5000 years ago in the Maghreb) that North Africa was incontestably marked by various cultural events. Then, Berbers experienced a long and complicated history with many invasions, conquests and migrations by Phoenicians, Romans, Vandals and Byzantines (Brett & Fentress 1996). The most significant event was the Arab conquest, begun during the 7th century, when North Africans were converted to Islam, and Arabic became the official unique language employed. In spite of strong resistance, Berbers acquiesced to Arab authority. Refractory groups were driven out and constrained to more isolated areas. This troubled past directly influenced the geographical distribution of Berber communities which are nowadays scattered in a vast region extending from Mauritania to Egypt (Siwa oasis) and from the Sahara desert to the Moroccan Atlas mountainous areas. Over the course of time, the various populations that migrated to North Africa have probably left a footprint in the gene pool of modern Berbers. 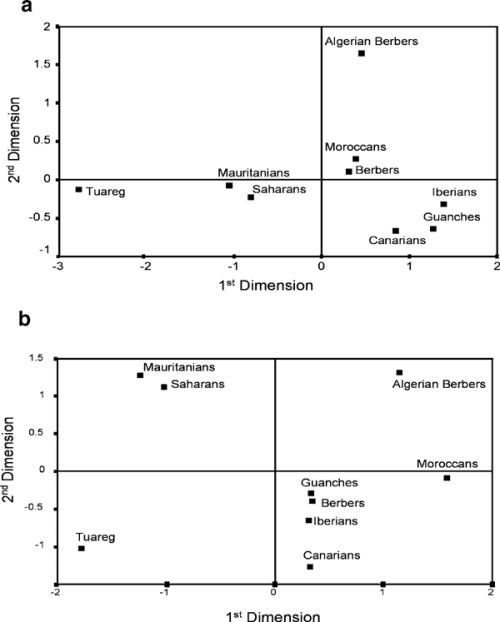 The genetic proximity observed between the Berbers and southern Europeans reveals that these groups shared a common ancestor. Two hypotheses are discussed: one would date these common origins in the Upper Paleolithic with the expansion of anatomically modern humans, from the Near East to both shores of the Mediterranean Sea; the other supports the Near Eastern origin, but would rather date it from the Neolithic, around 10,000 years ago (Ammerman & Cavalli-Sforza 1973; Barbujani et al. 1994; Myles et al. 2005; Rando et al. 1998). Common polymorphisms (i.e. those defining H and V lineages) between Berbers and south Europeans also could have been introduced or supported by genetic flows through the Straits of Gibraltar. For example, genetic exchanges could have taken place during prehistory, while European populations retreated from ice sheets and expanded from refuge, around 15,000 years ago (as evidenced by the H and U5b mitochondrial lineages). Alternatively, these exchanges could have occurred during history, with the invasion and the occupation during nearly seven centuries (from the 8th to the 15th century) of the Iberian Peninsula by Almoravide then Almohade Muslim Berber troops. 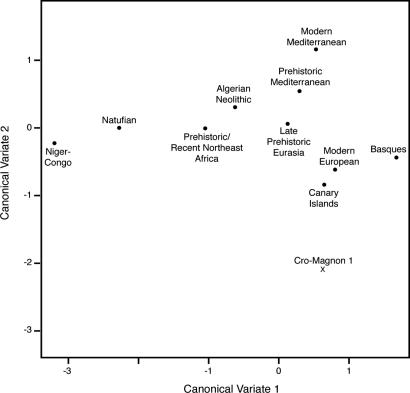 The differentiation observed between North Africans and sub-Saharan populations shows, first, that settlement of these areas was achieved by different migration waves and, then, that a genetic diversity was already observable in Africa since very old times. However, the Berber genetic heritage consists of a relatively high frequency of L lineages from various parts of Africa (i.e. L0a, L3i, L4, and L5 clades are from East Africa, L1b, L2b, and L3b are from West Africa, and L3e originated in the Sudan). It poses a question about the Sahara desert role in population movements and exchanges. It should be specified that the Sahara was not always a desert, because it also underwent enormous variation between wet and dry, offering green spaces favorable for human occupation and animal domestication (Aumassip et al. 1994; Said & Faure 1990). Thus, this is plausible that exchanges between African prehistoric populations took place; exchanges during which markers typical of sub-Saharan groups would have been introduced into the Berber gene pool. Contacts between North Africa and great sub-Saharan empires (such as those of Ghana, of Mali, or the Songhai Empire) are also reported by history during trans-Saharan trade of gold, salt and slaves. Overall, it is clear that neither the Strait of Gibraltar nor the Sahara desert appears to have forced the movements and exchanges between South Europeans, North Africans, and sub-Saharans. This interaction took place since prehistory in a human and genetic framework already diversified, and the maternal ancestors of the Berbers were able to exchange some markers with surrounding populations with different cultures and genetic pools. Influences from the Middle East and East Africa are marked in Siwa, while southwestern European influences are observed in the Maghreb. Although the origin of these Eurasian and sub-Saharan lineages in the mitochondrial pool of Berbers is still questionable (whether due to common ancestry or past and/or current gene flow), certainly they were not diluted by the many historical invasions and migrations, leaving a clear maternal footprint in the contemporary populations. We conclude that the origins and diversity of Berber populations are old and complex, and these communities bear genetic characteristics resulting from various events of gene flow with surrounding and migrating populations. Coudray, Clotilde, et al. " The complex and diversified mitochondrial gene pool of Berber populations." Annals of human genetics 73.2 (2009): 196-214.  Berbers live in groups scattered across North Africa whose origins and genetic relationships with their neighbours are not well established. The first hypervariable segment of the mitochondrial DNA (mtDNA) control region was sequenced in a total of 155 individuals from three Tunisian Berber groups and compared to other North Africans. The mtDNA lineages found belong to a common set of mtDNA haplogroups already described in North Africa. Besides the autochthonous North African U6 haplogroup, a group of L3 lineages characterized by the transition at position 16041 seems to be restricted to North Africans, suggesting that an expansion of this group of lineages took place around 10500 years ago in North Africa, and spread to neighbouring populations. Principal components and the coordinate analyses show that some Berber groups (the Tuareg, the Mozabite, and the Chenini-Douiret) are outliers within the North African genetic landscape. This outlier position is consistent with an isolation process followed by genetic drift in haplotype frequencies, and with the high heterogeneity displayed by Berbers compared to Arab samples as shown in the AMOVA. Despite this Berber heterogeneity, no significant differences were found between Berber and Arab samples, suggesting that the Arabization was mainly a cultural process rather than a demographic replacement. The cultural differentiation present in North Africa between Berber and Arab samples seems not to reflect genetic differences between both groups, as shown in the AMOVA analyses, and the MDS and PC analyses. If Arabs in Northern Africa were mostly descendants of Middle Eastern Arabs, the frequencies of haplogroups such as N, U1, U3, U7, and HV that are much more prevalent in the Middle East than elsewhere should be larger in N. African Arabs than in Berbers. However, the opposite is observed: these haplogroups add up to 5% in N. African Arabs but to 10% in Berbers. Drift in some of the more isolated Berber populations could explain this observation. The lack of differentiation between North African Arabs and Berbers has also been observed using other genetic markers such as classical markers (Bosch et al. 1997); autosomal STRs (Bosch et al. 2000), Alu insertion polymorphisms (Comas et al. 2000); and Y-chromosome lineages (Bosch et al. 2001). This pattern suggests that the Arabization of the area was mainly a cultural process, rather than a demographic replacement of the Berber populations that inhabited the region where the Arabic expansion took place. The present data has failed to confirm an east-west differentiation of North African populations as previously suggested using mtDNA sequences (Plaza et al. 2003) or other genetic markers (Bosch et al. 1997). The present mtDNA data show a more patchy genetic landscape in North Africa, with some Berber samples acting as outliers in the general North African landscape. The lack of mtDNA data for large geographic regions like the Kabylie (Algeria) and Libya, and the large number of isolated Berber samples considered in the present analysis may decrease the power to find the longitudinal differentiation previously shown by other studies. Fadhlaoui‐Zid, K., et al. " Mitochondrial DNA heterogeneity in Tunisian Berbers." Annals of human genetics 68.3 (2004): 222-233. |
|
|
|
Post by Admin on Dec 19, 2013 0:29:10 GMT
It's a busy time in our attempts to study our species' pre-modern history. Just two weeks ago, researchers reported the sequence of the oldest bones to yield human DNA. Now, the same research group is back with an entire genome, obtained from a bone found in Siberia's Denisova cave. This genome comes from a Neanderthal, but all the data reveals a lot about all the interconnections among the pre-modern human groups that were wandering around Eurasia tens of thousands of years ago. The analysis came with a tantalizing hint that one of those groups had interbred with a species separated from modern humans by over a million years—perhaps Homo erectus.  Excavations in the Denisovan cave have yielded tiny bone fragments that have had an outsized impact on our understanding of human evolution. The Denisova cave is famous for having yielded the bones that helped us identify the Denisovans, a group of archaic humans that inhabited Asia at the same time as the Neanderthals. Although we haven't found enough bones to know much about what the Denisovans looked like, DNA analysis has revealed that they are most closely related to Neanderthals and that they interbred with modern humans that went on to populate East Asia, the Americas, and the Pacific.  Inference of population size change over time. The y axis specifies a number proportional to the population size Ne. The x axis specifies time in units of divergence per base pair (along the top in years for mutation rates of 0.5 × 10−9 to 1.0 × 10−9 per site per year). Inference of population size change over time. The y axis specifies a number proportional to the population size Ne. The x axis specifies time in units of divergence per base pair (along the top in years for mutation rates of 0.5 × 10−9 to 1.0 × 10−9 per site per year).The new results spring from a toe bone found in the same cave, this one from a layer that is estimated to be tens of thousands of years earlier. DNA sequencing revealed the bone to be from a Neanderthal, a different group of pre-modern humans that is most closely related to the Denisovans. The DNA was in excellent condition and had a minimal (about one percent) contamination with sequences from modern humans. The team generated a high-quality genome using samples from this bone.  Relatedness of introgressing archaic and sequenced archaic samples. Divergence of phased present-day human genomes to archaic genomes in windows of size 0.01 cM with a minimum of 25,000 analysed bases. Relatedness of introgressing archaic and sequenced archaic samples. Divergence of phased present-day human genomes to archaic genomes in windows of size 0.01 cM with a minimum of 25,000 analysed bases.The sequence that resulted tells us a lot about Neanderthals. For one, it shows that other populations we've obtained DNA from (samples found in the Caucasus and Croatia) were closely related but distinct, indicating that the Neanderthals were already well established by the time this individual died. Those populations were apparently quite small, however, since there's not a lot of genetic diversity among them. In the case of the specific individual in the Denisovan cave, the lack of diversity was quite severe. Rather than carrying two distinct sets of chromosomes, large stretches of the two chromosomes were identical, indicating that they were inherited from a single individual in the recent past. The extent of this identity suggests that the parents of this individual were half-siblings, although other combinations (uncle-niece, aunt-nephew) would also produce a similar pattern. But the more significant results come from what this new sequence tells us about the other groups of humans present at the time, including modern humans. To begin with, it confirms the rough timing of the split between the ancestors of modern humans and the ancestors of Neanderthals and Denisovans, which took place about 550,000-600,000 years ago. The Neanderthals and Denisovans became a distinct population about 400,000 years ago.  Neanderthal gene flow into Siberian Denisovans. Neanderthal gene flow into Siberian Denisovans.We used the two high-coverage archaic genomes and a hidden Markov model (HMM) to identify regions of specifically Neanderthal and specifically Denisovan ancestry in 13 experimentally phased present-day human genomes. In the Sardinian and French genomes from Europe we find genomic regions of Neanderthal origin and few or no regions of Denisovan origin. In contrast, in the Han Chinese, the Dai in southern China, and the Karitiana and Mixe in the Americas, we find, in addition to regions of Neanderthal origin, regions that are consistent with being of Denisovan origin (Zscore54.3 excess relative to the Europeans) (Supplementary Information section 13), in agreement with previous analysis based on low-coverage archaic genomes. These regions are also more closely related to the Denisova genome than the few regions identified in Europeans. We estimate that the Denisovan contribution to mainland Asian and Native American populations is ,0.2% and thus about 25 times smaller than the Denisovan contribution to populations in Papua New Guinea and Australia. The failure to detect any larger Denisovan contribution in the genome of a 40,000-year-old modern human from the Beijing area suggests that any Denisovan contribution to modern humans in mainland Asia was always quantitatively small. In fact, we cannot, at the moment, exclude that the Denisovan contribution to people across mainland Asia is owing to gene flow from ancestors of present-day people in Oceania after they mixed with Denisovans. We also note that in addition to this Denisovan contribution, the genomes of the populations in Asia and America appear to contain more regions of Neanderthal origin than populations in Europe.  Altai and Denisovan allele sharing with Africans stratified by African allele frequency. The plot shows the D-statistic of the form D (Neanderthal, Denisova; Africa, chimpanzee) binned by derived allele count in 10 deeply sequenced African genomes. Altai and Denisovan allele sharing with Africans stratified by African allele frequency. The plot shows the D-statistic of the form D (Neanderthal, Denisova; Africa, chimpanzee) binned by derived allele count in 10 deeply sequenced African genomes.Our ancestors weren't the only ones who couldn't resist getting a piece of the Neanderthals. At least a half percent of the Denisovan genome also came from them as well. But perhaps the most unexpected finding comes from a comparison with the Denisovan genome. Modern humans in Africa never overlapped geographically with Neanderthals or Denisovans and thus contain none of their DNA. Therefore, any shared DNA they have should be inherited from a common ancestor, and the African's should be equally distant from the Neanderthals and Denisovans. Yet they're not. The Denisovans have some sequences that are much more distant than you'd expect.  Neanderthal gene flow into Siberian Denisovans. Neanderthal gene flow into Siberian Denisovans.After considering and rejecting a couple of alternative explanations for this, the paper settles on a rather radical explanation: Denisovans themselves interbred with a population that had been separated from their common ancestor with modern humans for about a million years. This, as the authors note, suggests that the DNA's source was Homo erectus. In fact, they suggest that the Denisovan's entire mitochondrial genome might have come from this interbreeding event, since it's much more distant from the Neanderthals' than the rest of the genome is. Of course, that explanation is even harder to square with the findings from the ancient bones in Spain, which had a similar sequence but came from skeletons that looked somewhat like Neanderthals. Unless, of course, the Spanish population also interbred with Homo erectus (or whatever this is) at some point. In any case, the results add yet another layer onto the increasingly complicated Out-of-Africa model of the origin of modern humans. We still arose in Africa and migrated out into Eurasia. But once we got there, we interbred with a previous wave of African expatriates and incorporated a small bit of their genetic legacy into our own. And one part of that previous wave may have even incorporated a tiny piece of a species that hadn't seen Africa for a very long time. Nature, 2013. DOI: 10.1038/nature12886 |
|
|
|
Post by Admin on Dec 23, 2013 21:21:26 GMT
The description of a Neanderthal hyoid from Kebara Cave (Israel) in 1989 fuelled scientific debate on the evolution of speech and complex language. Gross anatomy of the Kebara 2 hyoid differs little from that of modern humans. However, whether Homo neanderthalensis could use speech or complex language remains controversial. Similarity in overall shape does not necessarily demonstrate that the Kebara 2 hyoid was used in the same way as that of Homo sapiens. The mechanical performance of whole bones is partly controlled by internal trabecular geometries, regulated by bone-remodelling in response to the forces applied. Here we show that the Neanderthal and modern human hyoids also present very similar internal architectures and micro-biomechanical behaviours. Our study incorporates detailed analysis of histology, meticulous reconstruction of musculature, and computational biomechanical analysis with models incorporating internal micro-geometry. Because internal architecture reflects the loadings to which a bone is routinely subjected, our findings are consistent with a capacity for speech in the Neanderthals.  Figure 1. Male Homo sapiens and Pan troglodytes hyoid bones. Figure 1. Male Homo sapiens and Pan troglodytes hyoid bones.
Note that the human hyoid (A) lacks the large and distinctive bulla of the chimpanzee hyoid (B). Specimens are research quality casts numbers 844 and 837 held at the University Museum, Trieste.
The Kebara 2 Neanderthal dates from approximately 60 ka and is part of a near-complete adult male skeleton unearthed in 1983 [1]. Subsequent discoveries of additional fossil hominin hyoids have generated renewed interest in the bone’s potential to inform on the evolution of speech and complex language. These include: a partial Neanderthal hyoid (SDR-034) from El Sidròn Cave (Asturias, Spain) dated to ~43 ka [2]; two Middle Pleistocene hyoids (AT-1500 and AT-2000) assigned to Homo heidelbergensis from Sierra de Atapuerca (Spain) dated at ~530 ka [3]; and a “chimpanzee-like” hyoid assigned to Australopithecus afarensis from Dikika (Ethiopia, ~3.3 Ma) [4]. Gross anatomy of the hyoid in Pan troglodytes, which includes a cup-shaped extension or bulla (also present for the Dikika A. afarensis specimen), is very different to that of modern humans (Figure 1). However, analyses of gross macroscopic anatomy in the Kebara 2 hyoid (Figure 2A), as well as SDR-034, have shown that the hyoid of H. neanderthalensis was almost indistinguishable from that of modern humans [1], [2]. Similarly, anatomical and anthropometric descriptions of the Sima de los Huesos material show that the hyoid of H. heidelbergensis was modern-human-like [3]. Thus, it appears that the external macroscopic morphology of this important component of the vocal apparatus in modern humans had arisen by ~530 ka and has remained largely unchanged since. 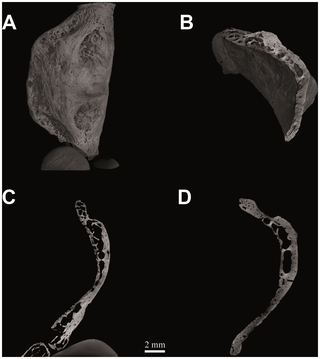 Figure 2. Figure 2. Computed tomography of Homo neanderthalensis (Kebara 2, Tel Aviv University - Israel). Figure 2. Figure 2. Computed tomography of Homo neanderthalensis (Kebara 2, Tel Aviv University - Israel).
Hyoid body volume rendering (V = 80 kV, I = 100 µA; pixel size: 10.0 µm; exposure time: 3.0 sec.; 2400 projections over 360 degrees) (a); spongy bone structure (b); histological architecture: medial sagittal section (c) and medial transverse section (d).Overall similarity between the external morphology of the Kebara 2 hyoid and those of modern humans has suggested to some researchers that the Kebara 2 Neanderthal was capable of speech, and perhaps language [1], [5]. Others have contested this conclusion, and, whether or not Neanderthals could speak remains a contentious issue [6]–[8]. Certainly a bone’s overall shape and external dimensions alone provide incomplete understanding of its precise function [9]. More detailed and specific insights into mechanical performance are reflected in the geometry of internal microstructure, including trabecular networks that are controlled through bone remodelling [9]–[11]. In remodelling, bone is resorbed or new ossification takes place, largely in response to mechanical loading. This is manifested in the specific morphology and orientation of the bony trabeculae and the size and distributions of the osteons [12], [13]. As observed in other fossil bones, histological structure reflects the forces imposed by muscles [14] and sound-waves during phonation [15]. Examination of internal microscopic anatomy reveals that the medial sagittal microCT section from the Kebara 2 hyoid body (Figure 2) shows a marked arcuate shape, corresponding with deep fossae for the insertion of the geniohyoid muscles [1]. Its histomorphology (Figure 2) is characterized by cortical bone with vascular channels, well-developed intertrabecular spaces and dorsoventrally oriented bony lamellae. In each of these respects its microarchitecture is comparable to that of modern human hyoids (Figure 3). 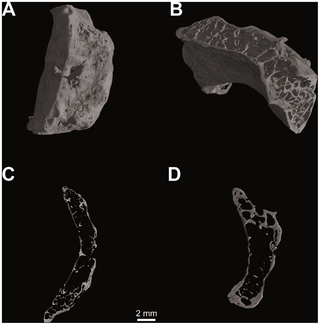 Figure 3. Computed tomography of Homo sapiens (N. S36-Sulmona Fonte d′Amore T64, University Museum Chieti - Italy). Figure 3. Computed tomography of Homo sapiens (N. S36-Sulmona Fonte d′Amore T64, University Museum Chieti - Italy).
Hyoid body volume rendering (V = 80 kV, I = 100 µA; pixel size: 12.5 µm; exposure time: 2.0 sec.; 2400 projections over 360 degrees) (a); spongy bone structure (b); histological architecture: medial sagittal section (c) and medial transverse section (d).Visual plots of von Mises (VM) stress distributions are given in Figure 4. VM stress is a good indicator of material failure in relatively ductile materials such as bone [22]. Mean values for VM stresses are given for each Finite Element Model (FEM) as a whole and for subgroups containing only surface elements in Table 1. Using the Graph Tool in Strand7 (2.4) a straight-line was drawn between the dorso-lateral-most extremes of each model to plot a graph of VM stress for elements intersecting the line (Figure 4). 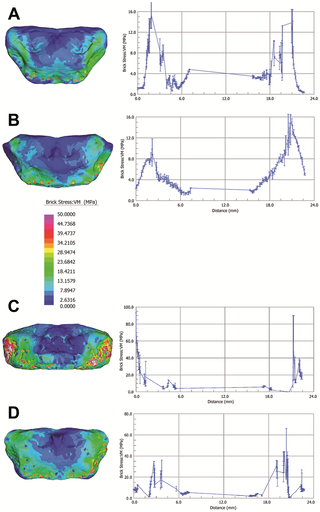 Figure 4. Computational biomechanical analyses of hyoid models. Figure 4. Computational biomechanical analyses of hyoid models.
Homo neanderthalensis (Kebara 2) (A), Homo sapiens (SAT37) (B), Homo sapiens (OP1T37) (C), and Homo sapiens (SAT41) (D). Surface von Mises stress distributions in visual plots for each model are given for each model on the left. On the right a two dimensional graph, generated using the Graph Tool (Vs Position) in Strand7 (2.4), is provided. This gives von Mises stress for internal elements intersecting a straight line drawn between nodes at maximum lateral width of each hyoid body, i.e., between the lateral most extremes where the body would have connected with the hyoid’s greater cornua (greater horns). Values are interpolated across element edges intersected by the line. MPa = megapascals.MicroCT analysis reveals that the hyoid bodies of both Kebara 2 and modern humans are characterized by two thick cortical layers, well-defined vascular channels and well-developed spongy structures. The detail of histological structure in all specimens, including Kebara 2, is typical of bone involved in intense and continuous metabolic activity. Our analysis shows that the similarity in gross surface morphology between the Kebara 2 hyoid and those of modern humans also extends internally to microscopic architecture and the orientations of the bony trabeculae comprising the spongy bone of the hyoid body. The results of FEA-based comparisons of our high-resolution models further show that the Kebara 2 hyoid presents very similar micro-biomechanical performance to that of modern humans under identical loadings. Minor histomorphological differences are present in that the Kebara 2 hyoid appears more dorsoventrally flattened in the distal regions, and the bony trabeculae appear thicker than in our modern human sample. However, given the considerable variation among the modern human specimens we consider it likely that these differences are a manifestation of individual histological variability and/or size differences. Modern-human-like gross anatomy in the hyoid body of a fossil specimen is not, in itself, clear demonstration that the individual was capable of speech [6]. However, our analyses demonstrate that previously observed gross similarities between the Kebara 2 hyoid and those of modern humans are not superficial. We conclude that the presence of modern-human-like histological features and micro-biomechanical behavior in the Kebara 2 hyoid indicates that this bone not only resembled that of a modern human, but that it was used in very similar ways. This is because the internal microarchitecture is a response to the vectors and magnitudes of the forces to which it is routinely subjected. These findings are consistent with the suggestion that the Kebara 2 Neanderthal practiced speech (sensu Duchin 1990) [29] although they do not prove that this was so. We are also mindful of the fact that our sample size is small and that the addition of further models of more modern human material, as well as specimens of Pan troglodytes and/or Pan paniscus, are needed before any firmer conclusions could be drawn. Micro-Biomechanics of the Kebara 2 Hyoid and Its Implications for Speech in Neanderthals |
|














