|
|
Post by Admin on Apr 11, 2016 1:54:44 GMT
 Figure 1 Tree Inference Previous research has shown that the DNA of modern humans is from 2.5 to 4 percent Neanderthal DNA, a legacy of breeding between modern humans and Neanderthals 50,000 years ago. As a result, the team was excited to find that, unlike other kinds of DNA, the Neanderthal Y chromosome DNA was apparently not passed to modern humans during this time. The Neanderthal Y chromosome genes could have simply drifted out of the human gene pool by chance over the millennia. Another possibility is that Neanderthal Y chromosomes include genes that are incompatible with other human genes. With the chimpanzee Y chromosome as the outgroup, three tree topologies could have related the lineages of the Neandertal, haplogroup A00, and the human reference (Figure 1A). To identify which of the three was consistent with the data, the key question was which of the three possible pairs of sequences is the most closely related. Of 118,643 sites (Figure 1B, filter 1) for which we had Neandertal data and human-chimpanzee reference alignments,18 we identified 24 biallelic SNVs for which the Neandertal sequence shared the chimpanzee allele and differed from both A00 and the human reference. In contrast, the chimpanzee and A00 sequences shared just four SNVs not present in the other sequences, and the chimpanzee and human reference sequences shared zero. Taken together, these data strongly support the tree that places the Neandertal Y as the most distantly related to the others (Figure 1A, tree i). Two of the four variants that are inconsistent with this topology are known to segregate within modern humans and are therefore the result of recurrent mutations or contamination (Appendix A).  Figure 2 Estimating the TMRCA of Neandertal and Modern Y Chromosomes We have estimated that the Neandertal Y chromosome from El Sidrón diverged from those of modern humans ∼590 kya, a value similar to TMRCA estimates for mtDNA sequences: 400 kya to 800 kya.11, 12 This time estimate and the genealogy we have inferred strongly support the notion that the most recent common ancestor of these Y chromosomes belonged to the population from which Neandertals and modern humans diverged, thereby refuting three alternative hypotheses. A priori, the Neandertal Y could have introgressed from a super-archaic population5 (Figure 3, scenario a), but this would have led to a far greater TMRCA estimate. Alternatively, it could have introgressed from the ancestors of modern humans after their divergence from Neandertals and prior to the most recent common ancestor of present-day Y chromosomes (scenario b) or from modern human populations subsequent to their migrations out of Africa (scenario c). We can also reject these hypotheses, as each requires a more recent split time. The fact that the Neandertal Y-chromosome lineage we describe has never been observed in modern humans suggests that the lineage is most likely extinct. Although the Neandertal Y chromosome (and mtDNA) might have simply drifted out of the modern human gene pool,24 it is also possible that genetic incompatibilities contributed to their loss. In comparing the Neandertal lineage to those of modern humans, we identified four coding differences with predicted functional impacts, three missense and one nonsense (Table 1). Three mutations—within PCDH11Y, USP9Y, and TMSB4Y—are unique to the Neandertal lineage, and one, within KMD5D, is fixed in modern human sequences. The first gene, PCDH11Y, resides in the X-transposed region of the Y chromosome. Together with its X-chromosome homolog PCDH11X, it might play a role in brain lateralization and language development.25 The second gene, USP9Y, has been linked to ubiquitin-specific protease activity26 and might influence spermatogenesis.27 Expression of the third gene, TMSB4Y, might reduce cell proliferation in tumor cells, suggesting tumor suppressor function.28 Finally, the fourth gene, KDM5D, encodes a lysine-specific demethylase whose activity suppresses the invasiveness of some cancers.29  Figure 3 Relationship of Neandertal Y Chromosome to Those of Modern Humans Polypeptides from several Y-chromosome genes act as male-specific minor histocompatibility (H-Y) antigens that can elicit a maternal immune response during gestation. Such effects could be important drivers of secondary recurrent miscarriages30 and might play a role in the fraternal birth order effect of male sexual orientation.31 Interestingly, all three genes with potentially functional missense differences between the Neandertal and modern humans sequences are H-Y genes, including KDM5D, the first H-Y gene characterized.32 It is tempting to speculate that some of these mutations might have led to genetic incompatibilities between modern humans and Neandertals and to the consequent loss of Neandertal Y chromosomes in modern human populations. Indeed, reduced fertility or viability of hybrid offspring with Neandertal Y chromosomes is fully consistent with Haldane’s rule, which states that “when in the [first generation] offspring of two different animal races one sex is absent, rare, or sterile, that sex is the [heterogametic] sex.”33 DOI: dx.doi.org/10.1016/j.ajhg.2016.02.023 |
|
|
|
Post by Admin on Apr 13, 2016 1:17:16 GMT
There is evidence that our ancestors interbred with Neanderthals and exchanged genes associated with disease. There is also evidence that viruses moved into humans from other hominins while still in Africa. So, the researchers argue, it makes sense to assume that humans could, in turn, pass disease to Neanderthals, and that -- if we were mating with them -- we probably did. Dr Charlotte Houldcroft, from Cambridge's Division of Biological Anthropology, says that many of the infections likely to have passed from humans to Neanderthals -- such as tapeworm, tuberculosis, stomach ulcers and types of herpes -- are chronic diseases that would have weakened the hunter-gathering Neanderthals, making them less fit and able to find food, which could have catalysed extinction of the species.  "Humans migrating out of Africa would have been a significant reservoir of tropical diseases," says Houldcroft. "For the Neanderthal population of Eurasia, adapted to that geographical infectious disease environment, exposure to new pathogens carried out of Africa may have been catastrophic." "However, it is unlikely to have been similar to Columbus bringing disease into America and decimating native populations. It's more likely that small bands of Neanderthals each had their own infection disasters, weakening the group and tipping the balance against survival," says Houldcroft.  The researchers describe Helicobacter pylori, a bacterium that causes stomach ulcers, as a prime candidate for a disease that humans may have passed to Neanderthals. It is estimated to have first infected humans in Africa 88 to 116 thousand years ago, and arrived in Europe after 52,000 years ago. The most recent evidence suggests Neanderthals died out around 40,000 years ago. Another candidate is herpes simplex 2, the virus which causes genital herpes. There is evidence preserved in the genome of this disease that suggests it was transmitted to humans in Africa 1.6 million years ago from another, currently unknown hominin species that in turn acquired it from chimpanzees. |
|
|
|
Post by Admin on Apr 24, 2016 22:48:48 GMT
 Analysis of dental calculus is increasingly important in archaeology, although the focus has hitherto been on dietary reconstruction. Non-edible material has, however, recently been extracted from the dental calculus of a Neanderthal population from the 49 000-year-old site of El Sidrón, Spain, in the form of fibre and chemical compounds that indicate conifer wood. Associated dental wear confirms that the teeth were being used for non-dietary activities. These results highlight the importance of dental calculus as a source of wider biographical information, and demonstrate the need to include associated data within research, in particular tooth wear, to maximise this valuable resource. A new study, published in the journal Antiquity, has discovered evidence of non-edible material in Neanderthal teeth. It is a remarkable find, one which opens up a host of possibilities concerning what Neanderthals did with their teeth: did they use their mouths as a third hand, or are the non-edible materials evidence that Neanderthals were concerned with dental hygiene? 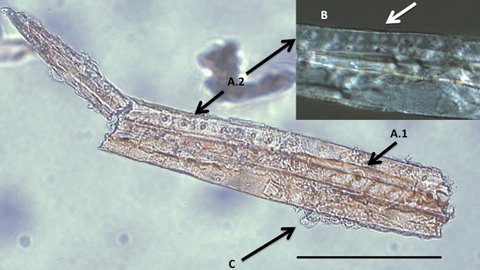 Most estimates suggest that Neanderthals first came on the scene between 300,000 and 250,000 years ago. By around 32,000 years ago they had been rendered completely extinct. They are often portrayed as being less intelligent, and uncivilised when compared to humans, however, new studies, including this latest one by Anita Radinia, Stephen Buckleya, Antonio Rosasa, Almudena Estalrricha, Marco de la Rasillaa and Karen Hardy; are increasingly bringing this assumption into question. For their research, the team led by Hardy from the Catalan Institution for Research and Advanced Studies extracted chemical compounds and micro fossils embedded in the teeth of Neanderthals from the 49,000 year old El Sidrón site in Spain. It was a unique approach to the still relatively new use of dental calculus (tartar) in archaeological study.  Hardy’s team discovered traces of conifer wood from a non-edible part of the trees in the molar of one adult Neanderthal. According to the study, the substance with no nutritional value could not have been confused with an edible part of a conifer – meaning it’s presence was certainly not connected to the Neanderthal’s diet. The study, published in the April edition of Antiquity, points out that associated dental wear suggests the teeth were being used for non-dietary functions, supporting the idea the piece of conifer had become embedded in the molar because the animal was using its mouth as a third hand. However, there are several other possible explanations. DOI: dx.doi.org/10.15184/aqy.2016.21 |
|
|
|
Post by Admin on Apr 28, 2016 22:49:41 GMT
 To assess whether the ancestors of the Oase 1 individual mixed with Neanderthals, we tested whether the Altai Neanderthal genome shares more alleles with the Oase 1 genome than with sub-Saharan Africans. We find this to be the case (|Z|=7.7; Supplementary Information section 4). We then asked if the amount of Neanderthal ancestry in the Oase 1 genome is similar to that in present-day non-Africans. Surprisingly, the Neanderthal genome shares more alleles with the Oase 1 individual than it does with any present-day people in Eurasia that we tested indicating that he carries more Neanderthal-like DNA than present-day people (5.0≤|Z|≤8.2; Extended Data Table 3). We also observe more Neanderthal-like alleles in the Oase 1 individual when we compare him to four early modern humans: an 8,000-year-old individual from Luxembourg, and three individuals from Russia who vary in age between 24,000 and 45,000 years (3.6≤|Z|≤6.8; Extended Data Table 3). Thus, the Oase 1 individual appears to have carried more Neanderthal-like DNA than any other modern human analyzed to date. This observation cannot be explained by residual present-day human contamination among the DNA fragments that carry terminal C to T substitutions, because all modern humans studied to date carry less Neanderthal ancestry than the Oase 1 genome, and thus contamination would lower, rather than increase, the apparent Neanderthal ancestry. We estimated the proportion of Neanderthal DNA in the Oase 1 genome using three different statistics7,29 (Supplementary Information section 4). Although the results differ, they all yield point estimates between 6.0% and 9.4% (Table 1). For one of the statistics, none of the 90% confidence intervals for Neanderthal ancestry in the other modern human samples overlap with the confidence interval in Oase 1. When we restrict analysis to transversion SNPs, the point estimates of Neanderthal ancestry are even higher (range of 8.4% to 11.3%) (Extended Data Table 4).  Figure 1 Allele sharing between the Oase 1 individual and other genomes To study the spatial distribution of Neanderthal DNA across the Oase 1 genome, we designed capture probes for around 1.7 million nucleotide positions where nearly all individuals in a sub-Saharan African population carry one allele while Neanderthal genomes carry another allele. We used these probes to isolate DNA fragments from the Oase 1 individual. A total of 78,055 sites were covered by deaminated DNA fragments from the Oase 1 individual as well as from the ~36,000- to 39,000-year-old Kostenki 14 individual from western Russia16, the ~43,000- to 47,000-year-old individual from Ust’-Ishim in Siberia15, and three present-day human genomes from China, France and Sudan (Supplementary Information section 5). Because the Dinka from Sudan are thought to have little or no Neanderthal ancestry7, we subtracted the number of alleles that match the Neanderthals in the Dinka individual (485) from the number in the other genomes to estimate the number of alleles attributable to Neanderthal ancestry. The resulting numbers of putative Neanderthal alleles are 3,746 in the Oase 1 individual, 1,586 and 1,121 in the Ust’-Ishim and Kostenki 14 individuals, respectively, and 1,322 and 1,033 in the Chinese and the European individuals (Extended Data Table 5). Thus, the Neanderthal contribution to the Oase 1 genome appears to be between 2.3- and 3.6-fold larger than to the other genomes analyzed. Assuming that the Neanderthal contribution to the European individual is 2%7, this suggests that 7.3% of the Oase 1 genome is of Neanderthal origin. When the numbers of alleles matching the Neanderthal genome are compared per chromosome (Extended Data Table 5), the highest numbers are always observed for the Oase 1 genome, except in the case of chromosome 21, where the Ust’-Ishim individual carries a large segment of likely Neanderthal ancestry. We plotted the positions of Neanderthal-like alleles across the Oase 1 genome (Fig. 2). We detect three segments that are over 50 cM in size, suggesting that the Neanderthal contribution to the Oase 1 individual occurred so recently in his family tree that chromosomal segments of Neanderthal origin had little time to break up due to recombination. To estimate the date of the most recent Neanderthal contribution to the Oase 1 genome, we studied the size spans of seven segments of the genome that we could clearly identify as being recently derived from Neanderthals. Their genetic lengths suggest that the Oase 1 individual had a Neanderthal ancestor as a 4th, 5th, or 6th degree relative (Supplementary Information section 5). This would predict that an average of 1.6% to 12.5% of the Oase 1 genome derived from this Neanderthal ancestor, which is in the range of our Neanderthal ancestry estimates (Extended Data Table 5). Visual inspection of the Oase 1 genome suggests that in addition to these seven segments, other smaller segments also carry Neanderthal-like alleles (Fig. 2). When we remove the seven longest segments, the estimate of Neanderthal ancestry in Oase 1 drops from 7.3% to 4.8%, which is still around twice the 2.0%–2.9% estimated for the French, Han, Kostenki and Ust’-Ishim individuals in this remaining part of the genome. This additional Neanderthal ancestry could reflect an older Neanderthal admixture into the ancestors of Oase 1, or that we failed to find all segments of recent Neanderthal ancestry.  Table 1 Estimated fraction of the Oase 1 genome that derives from Neanderthals. The Oase 1 genome shows that mixture between modern humans and Neanderthals was not limited to the first ancestors of present-day people to leave Africa, nor to the Near East; it occurred later as well and probably in Europe. The fact that the Oase 1 individual had a Neanderthal ancestor removed by only four to six generations allows this Neanderthal admixture to be dated to less than 200 years before the time he lived. However, the absence of a clear relationship of the Oase 1 individual to later modern humans in Europe suggests that he may have been a member of an initial early modern human population that interbred with Neanderthals but did not contribute much to later European populations. To better understand the interactions between early modern and Neanderthal populations, it will be important to study other specimens that, like Oase 1, have been suggested to carry morphological traits suggestive of admixture with Neanderthals30. At each SNP covered at least once in Oase 1, we selected the majority allele (in case of a tie, we picked a random allele). We then merged the Oase 1 data with 25 genomes of present-day humans sequenced to 24–42× coverage7, the Altai Neanderthal7, the Siberian Denisovan9, a ~45,000-year-old modern human from Ust’-Ishim in Siberia15, an ~8,000-year-old Mesolithic individual from Loschbour Cave, Luxembourg28 and a ~7,000-year-old early farmer from Stuttgart, Germany28 (Extended Data Table 9). All the calls for the five deeply sequenced ancient genomes were performed in the same way. We restricted analyses to sites with a minimum root-mean-square mapping quality of 30 in the 30 genomes. We added lower coverage shotgun data from the ~36,000-year-old Kostenki 14 from Russia16, the ~24,000-year-old Mal’ta Siberian individual from Russia40, an 8,000-year-old Mesolithic individual from La Brana Cave, Spain41, a Neanderthal from Mezmaiskaya in Russia7, and a pool of three Neanderthals from Vindija Cave in Croatia6, in these cases restricting to fragments with MAPQ≥37 to match the filter for the low coverage Oase 1 data (Extended Data Table 9). Nature. 2015 Aug 13; 524(7564): 216–219. |
|
|
|
Post by Admin on May 19, 2016 22:47:10 GMT
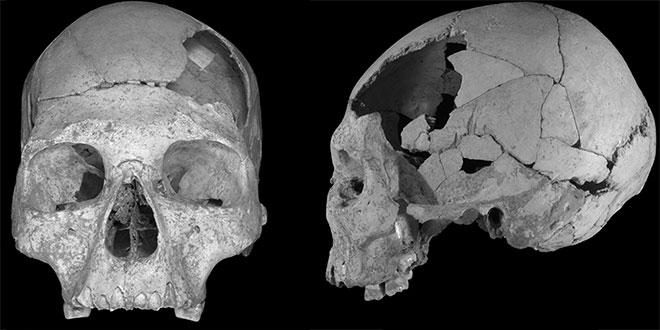
 Last Wednesday Papagianni, a researcher from the Centre for Archaeology of Human Origins at the University of Southampton, gave the inaugural seminar for the newly minted Center for Climate and Life at Columbia University and the Lamont-Doherty Earth Observatory. In her talk, titled "The Neanderthal Paradox," she explained the differences between the Neanderthals, a species that went extinct, and the ancient humans, our tropical-adapted forebears that would defy all odds and usurp Neanderthals as they migrated from Africa and into the chilly north. It was a talk that wove together culture, climate change, genetics and evolution. Overall, it was an apt introduction to usher in this new division of Climate and Life. Though they were closely related, Neanderthals and ancient humans were different species. Papagianni compared the skull of a Neanderthal to that of a human. The difference is clear. "Think of the skull of a human as a soccer ball, and the skull of a Neanderthal as a football," she says. This sports analogy goes a step further. Like a soccer player, humans are lean and well-suited to running. Neanderthals have stocky statures and barrel chests, more like the stereotypical football player. Because of this, Neanderthals were better adapted to colder weather than ancient humans who originated in the tropical climates of Africa.  "What you likely know about Neanderthals," Papagianni says, "it's that they're supposed to be stupid." This is a tough rap, but archaeological research suggests they were advanced enough to make clothes and build fires, which would have been critical to survive the cold weather. They also used stone tools. Additionally, based on the size of their skulls, the brains housed with them were likely large enough to warrant some form of language capability. This language hypothesis is supported by modern experiments that show learning how to make and wield stone tools required some form of verbal instruction.  Humans had diets higher in energy-rich meat that could support smaller stomachs and bigger brains. They invented tools with multiple uses that could adapt to different circumstances. They had more advanced language capabilities to pass on these skills. They were smarter; one could almost say they had more culture. "And if you're smart and you can speak," Papagianni quips, "you want to go to Europe." So the humans migrated north. This is where the genetic factors come into play. Neanderthal communities became fractured. As they grew more and more isolated, their gene pool evaporated into a spattering of puddles. This so called genetic bottleneck can lead to the demise of a species when genetic diversity gets prohibitively low.  Papagianni explained the current theories for how humans were able to persist in the North at the expense of Neanderthals, but she ended on an ongoing research question: Why did humans leave Africa in the first place? Their migration could have been sparked by competition, climate change or simply a great hallmark of human nature, curiosity. Over the past 2 million years, humans have proven to be a remarkably successful species. In fact, humans are the only species on the planet with a global distribution. In order to figure out how we might fare in the future with a changing planet, it's critical to get insight from our past. The research of Papagianni, as well as the new Center for Climate and Life, will yield this essential insight into Earth's past and future, and our place within it. |
|














