|
|
Post by Admin on Oct 18, 2016 20:57:14 GMT
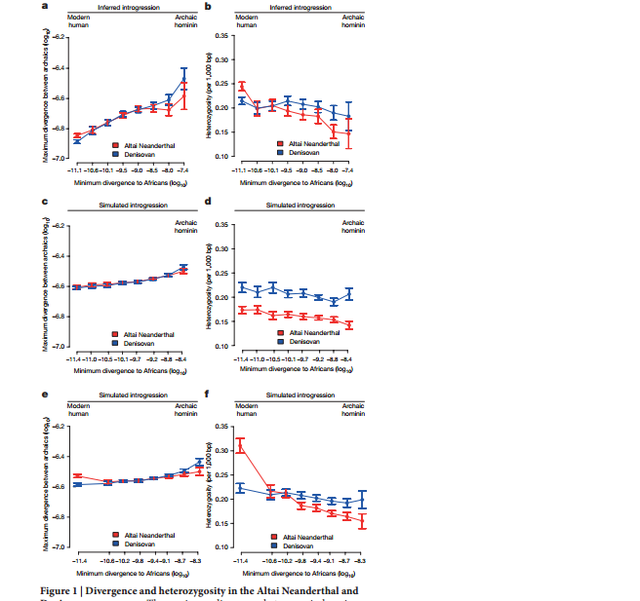 Figure 1 The Altai Neanderthal genome shares 5.4% more derived alleles with present-day Africans than does the Denisovan genome. This excess is particularly pronounced for derived alleles found at >0.9 frequency in Africans (Extended Data Table 1). These observations have been interpreted as evidence of gene flow from an unknown and more deeply diverged archaic hominin into the Denisovan lineage2 . Here we examine whether gene flow from modern humans into the ancestors of the Altai Neanderthal may also have occurred. Noting that regions in the Denisovan genome introgressed from a deeply divergent archaic hominin should have unusually high divergence to present-day Africans, and that regions of the Altai Neanderthal genome introgressed from modern humans should have unusually low divergence to them, we examined the divergence of these archaic genomes to 504 African genomes8 in 15,881 sequence windows of 100 kb (Supplementary Information section 9). 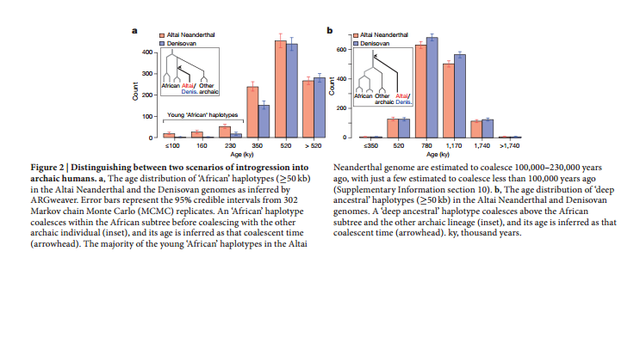 Figure 2 The inferred demographic model confirms and provides quantitative estimates of previously inferred gene flow events among modern and archaic humans2,3 (Extended Data Fig. 1). These include Neanderthal gene flow into modern humans outside Africa (3.3–5.8%) and gene flow from an unknown archaic hominin into the ancestors of Denisovans (0.0–0.5%). Interestingly, we also detect a signal of gene flow from modern humans into the ancestors of the Altai Neanderthal (1.0–7.1%). The precise source of this gene flow is unclear, but it appears to come from a population that either split from the ancestors of all present-day Africans or from one of the early African lineages, as significant admixture rates are estimated from San as well as Yoruba individuals. This introgression thus occurred in the opposite direction from the previously reported gene flow from Neanderthals to modern humans outside Africa2,3,12. Using parameters compatible with this model, we simulated windows of 100 kb for a model with gene flow into the Denisovan lineage from both the Altai Neanderthal and a deeply divergent archaic hominin2 , and a model including these admixture events together with modern human gene flow into the Altai Neanderthal lineage. Both models reproduced the observed patterns in windows most divergent to Africans (Fig. 1c and d), but only the model with modern human gene flow into the ancestors of the Altai Neanderthal reproduced the observed divergence and heterozygosity patterns in windows of the Altai Neanderthal least divergent to Africans (Fig. 1e and f). 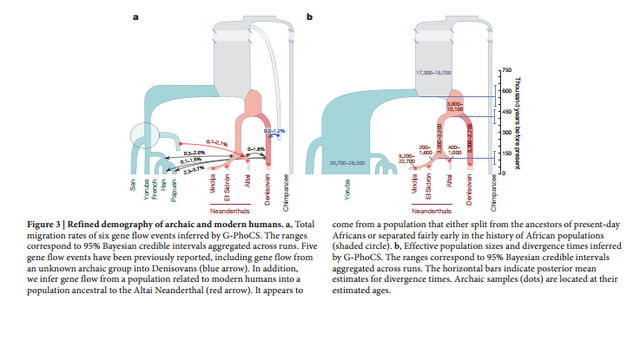 Figure 3 We find that the chromosome 21 of the Altai Neanderthal shares more derived alleles with Africans than the chromosome 21 of El Sidrón (3.5% more) and Vindija (4.9% more) Neanderthals, with the European Neanderthals sharing more derived alleles with Africans than the chromosome 21 of the Denisovan (9.8% more for El Sidrón, 8.8% more for Vindija). A comparison of the distribution of haplotype ages is not possible with the European Neanderthals, owing to insufficient amounts of data, but we compared the cumulative length of haplotypes coalescing within the African subtree for each Neanderthal lineage. |
|
|
|
Post by Admin on Nov 3, 2016 20:51:58 GMT
 Neanderthals in cold regions probably ate a lot more vegetable food than was previously thought. This is what archaeologist Robert Power has discovered based on new research on ancient Neanderthal dental plaque. PhD defence 1 November. Mammoth steppe Plants were an important part of the menu of Neanderthals who lived in the warmer Mediterranean regions of Eurasia between 180,000 and 30,000 years ago. But paleoanthropologists had for a long time assumed that the same did not apply in colder regions such as the Mammoth steppes. The Mammoth steppe, a region of steppe tundra almost completely devoid of trees, was the dominant landscape from Central Europe to East Asia during the cold periods of the Pleistocene era. Neanderthals in these areas were thought to have been carnivores, eating virtually only the flesh of large wild animals. This very limited diet made this hominid species vulnerable and may well have contributed to their becoming extinct, anthropologists reasoned.  Plant consumption Leiden archaeologist Robert Power discovered that Neanderthals must have consumed regularly plants as food even in this cold and dry environment. ‘The Mammoth steppe is an environment that we don’t really understand because it no longer exists due to climate change and megafauna extinction. It may well be that these ancient grasslands were far more useful to Neanderthals than we thought.’ New research methods The lack of comprehensive methods for tracing food consumptions is the reason why researchers have difficulty in establishing ancient diets. Thanks to emerging microscopy methods, plaque has become a good source of traces of food that can now be analysed. Power studied microbotanical particles in the plaque of Neanderthals from six different archaeological sites, including Croatia, Italy and Russia. His results from 48 teeth found that in all regions Neanderthals regularly ate plant food.  Neanderthals have unique pattern Power: ‘One of the big lessons of my work on Neanderthals was that the only reason we find the result surprising is that we expect Neanderthals to resemble modern human hunter gatherers in the way they foraged for food, but they didn’t. In the past we saw them as primitive cavemen with very basic behaviour, but now we have updated our view and see them as very modern. But that doesn’t mean we can afford to pigeonhole Neanderthals into our own particular version of humanity. They had their own unique pattern.’ |
|
|
|
Post by Admin on Nov 7, 2016 20:46:21 GMT
 In 2006, when the first two papers analyzing fragments of Neanderthal nuclear DNA sequence were published (Green et al., 2006, Noonan et al., 2006), the discordance between their conclusions suggested that contamination of the samples with modern human DNA remained a serious concern (Wall and Kim, 2007). The current paper soundly demonstrates that the effects of contamination can be brought under control, at least for mtDNA, and that, provided the sequencing can go deep enough in a DNA library of sufficient complexity, the prospects for convincingly determining the true Neanderthal sequence are excellent. The only caveat is that if errors or contaminants were more common than the true Neanderthal sequence, deep sequencing would only find the erroneous consensus. By sequencing to a 35× depth, the current paper not only obtains the desired sequence but also quantifies many aspects of the errors induced by DNA degradation.  Although the mtDNA data suggest that there was no admixing between Neanderthal and modern humans, this conclusion remains controversial. The estimated time of common ancestry between this Neanderthal and modern human mtDNA was 660,000 years ago (Figure 1). Yet there are many nuclear genes in the modern human genome where the best estimates of the time to most recent common ancestry are in the range of 1 to 4 million years ago, leaving open the possibility of admixture with even more ancient hominids (Garrigan and Hammer, 2006). The long period of coexistence of modern humans and Neanderthals, as well as the great depth of common ancestry of modern human nuclear genes, make it quite plausible that there was opportunity for interbreeding (Figure 1). If there had been admixture, say 100,000 years ago, giving modern humans small segregating pieces of our genome with Neanderthal ancestry, it would be nearly impossible to identify them as such, even with full genome sequences.  Whether the sum of the data lies in favor of some admixture or zero admixture remains unclear (Plagnol and Wall, 2006, Fagundes et al., 2007). The observation that mtDNA sequence from Neanderthals lies outside the range of modern human variation was made some years ago (Krings et al., 1997), and the conclusion then was that this was evidence against admixture. But Nordborg (1998)) pointed out that mtDNA follows clonal haploid transmission, and so the genealogy inferred from mtDNA is only one sampling among millions of possible genealogies. Admixture could have easily occurred without leaving any trace in current mtDNA sequences. Moreover, because the mtDNA data are from a single Neanderthal, it remains possible that other samples may produce a different conclusion, especially regarding the time of common ancestry. Also, perhaps the interbreeding was strictly unidirectional; for example, only human female by Neanderthal male matings occurred and never the reverse. This would yield modern humans with admixed nuclear genes but a complete absence of Neanderthal mtDNA. Although the data do not yet allow one to conclude that there was absolutely zero interbreeding, they nevertheless suggest that it was not rampant.  Natural selection is expected to perturb the background pattern of DNA sequence changes differently from the random sampling process of neutral drift, and we now have a plethora of statistical methods for inferring the signature of selection. The Neanderthal mtDNA sequence shows a clear elevation of nonsynonymous (amino-acid altering) changes relative to modern human polymorphisms, a pattern of change that is consistent with some degree of relaxed efficacy of purifying selection in Neanderthals. Green et al. argue that the most likely cause for this pattern is a smaller population size of Neanderthals than that of modern humans, making selection less efficient at removing slightly deleterious polymorphisms from the Neanderthal population. Positive adaptive selection would also accelerate nonsynonymous changes, but the data are strongly consistent with an overall pattern of purifying selection instead. The mitochondrial gene COX2 encoding subunit 2 of cytochrome c oxidase has four amino acid differences between modern human and Neanderthal. But even this substantial change did not attain significance for positive selection. Given the mere 0.5% sequence divergence between modern human and Neanderthal nuclear genes (Noonan et al., 2006), adaptive changes would have to generate a large sequence discrepancy to be detectable. The production of the complete mtDNA genome sequence of a Neanderthal sample is a grand success, and it whets the appetite for continuing the endeavor to produce a full nuclear genome sequence. Several attributes of the nuclear genome will make this a much less tractable challenge, including its sheer size, its redundancy and heterozygosity, an unknown error model (which will likely differ from that of mtDNA), and the challenge of identifying contaminants when the true divergence is likely only to be 0.5%. But one clear lesson from the new Green et al. study is that brute-force sequence depth can go far to resolve ambiguities and to distinguish the error from the true sequence. Perhaps the most informative attribute of the genetic data for determining whether or not there was human-Neanderthal admixture will come from quantifying patterns of shared polymorphisms between modern humans and Neanderthals. Such population genetic inference would require analysis of multiple Neanderthal samples, so it is fortunate that fossil remains of more than 400 of these beings have been found. |
|
|
|
Post by Admin on Nov 9, 2016 20:46:27 GMT
 In 2010 scientists compared a draft of the Neanderthal genome with that of modern humans and concluded that most humans have 1-4 per cent Neanderthal DNA. This could be explained either by interbreeding between Neanderthals and modern humans or because the two species share a common ancestor. Homo neanderthalensis lived during the last Ice Age and their bodies were adapted by natural selection to accommodate these harsh conditions. Neanderthal bodies were shorter and stockier than ours. Their skulls had a sloping forehead, distinct brow ridges, a large middle face, angled cheekbones and a huge nose adapted to humidify and warm the incoming cold, dry air. Neanderthal brains were at least as large as ours. Neanderthals ate plants but also considerable amounts of meat, because availability of plants dropped significantly during the extreme winters. They were specialised seasonal game hunters using sharp wooden spears, and their fossil bones show a high frequency of fractures similar to professional rodeo riders who regularly tangle with dangerous animals.  Powerful brutes? Neanderthals were long considered to be powerful, brutish people, able hunters but too primitive to show modern human behaviour. However, findings since the 1950s have challenged this view. In 1957 the remains of eight adults and two infants were discovered in northern Iraq and appear to have been purposefully buried, perhaps accompanied with flowers. Also, examination of the adult skeletons indicated that injuries had been tended and healed, suggesting Neanderthals cared for their sick and wounded. There is also evidence that Neanderthals used a diverse range of stone tools, used fire, made and wore clothes, made music and appreciated the medicinal qualities of some plants. Why did Neanderthals suddenly die out? Neanderthals and modern humans may have had little direct contact over tens of thousands of years until, during a particularly cold period, modern humans spread widely across Europe.  This may have prevented Neanderthals from returning to areas they once favoured, putting them on a steep, slippery slope towards extinction. Neanderthal numbers declined to the point of extinction over a few thousand years after modern humans moved into Eurasia, and all traces had vanished by about 40,000 years ago. Some propose that modern humans killed off the Neanderthals. This doesn’t seem that likely to me, because it is clear that Neanderthals were formidable hunters and would not be so easily wiped out in battles. The “make love not war” theory proposes that they were assimilated into the much larger modern human stock by interbreeding. Other theories as to why Neanderthals suddenly disappeared invoke climate change or dietary deficiencies, but nobody knows for sure. |
|
|
|
Post by Admin on Nov 19, 2016 20:53:23 GMT
Now, two new studies published in the journal Cell, help us to understand more about these differences. African immune systems are by and large stronger than their European counterparts, researchers found. Some of these variations can be traced back on the genetic level to proto-Europeans interbreeding with Neanderthals, which weakened their immune systems, over Africans who did not. Homo sapiens left Africa somewhere between 100,000 and 60,000 years ago, and encountered a Europe already colonized by Neanderthals. Eventually, the two interbred, and the immune responses of Neanderthals helped home sapiens adapt to their new environment and the pathogens there. Sometime after, around 40,000 years ago, Neanderthals vanished from the Earth. Today 20% of the world population carries Neanderthal genes within them.  Since Europe had a colder climate, a subtler inflammatory response was sufficient, researchers believe. However, in Africa, pathogens are more robust and so a faster immune response was required to ensure survival. Though they may respond better to pathogens, having an African immune system does have a downside, a higher risk of an autoimmune disorders. Lluis Quintana-Murci of Institut Pasteur and CNRS in Paris, led one study. He said their findings show that such differences are responses that have been transcribed into DNA. Since the single gene they discovered was passed on by Neanderthals already living in Europe, it is considered something that conveyed an evolutionary advantage.  RNA sequencing was used by Quintana-Murci and his team, to categorize immune cells called primary monocytes, taken from 200 participants, half of European and the other African ancestry. Researchers looked at how the cells would respond when encountering a certain bacteria or virus. Differences in gene activity within immune cells varied widely between populations. What differentiated these two studies from others is not only studying immune response but the gene expression behind them. Quintana-Murci and colleagues found that changes in a single gene, adopted from Neanderthals, became integral in the modern day European immune system and how it responds to pathogens. This discovery offers substantial evidence pertaining to gene selection and immune response. Certain regulatory variants too came from Neanderthals, which Europeans “borrowed” when the two interbred. This all plays out in how a Caucasian immune system responds to viral infections.  Luis Barreiro was the senior author of the other study. Barreiro hails from University of Montreal and the CHU Sainte-Justine, also in Canada. He and his team tested how African and European immune systems, particularly cells known as primary macrophages, responded to live, pathogenic bacteria. The macrophages of 175 Americans, 80 African Americans and 95 Caucasians, were used. These cells were grown in dishes and then infected with either listeria or salmonella bacteria. Summary Individuals from different populations vary considerably in their susceptibility to immune-related diseases. To understand how genetic variation and natural selection contribute to these differences, we tested for the effects of African versus European ancestry on the transcriptional response of primary macrophages to live bacterial pathogens. A total of 9.3% of macrophage-expressed genes show ancestry-associated differences in the gene regulatory response to infection, and African ancestry specifically predicts a stronger inflammatory response and reduced intracellular bacterial growth. A large proportion of these differences are under genetic control: for 804 genes, more than 75% of ancestry effects on the immune response can be explained by a single cis- or trans-acting expression quantitative trait locus (eQTL). Finally, we show that genetic effects on the immune response are strongly enriched for recent, population-specific signatures of adaptation. Together, our results demonstrate how historical selective events continue to shape human phenotypic diversity today, including for traits that are key to controlling infection. |
|
















