|
|
Post by Admin on Apr 13, 2015 13:56:38 GMT
 The calcite-encrusted skeleton of an ancient human, still embedded in rock deep inside a cave in Italy, has yielded the oldest Neanderthal DNA ever found. These molecules, which could be up to 170,000 years old, could one day help yield the most complete picture yet of Neanderthal life, researchers say. Although modern humans are the only remaining human lineage, many others once lived on Earth. The closest extinct relatives of modern humans were the Neanderthals, who lived in Europe and Asia until they went extinct about 40,000 years ago. Recent findings revealed that Neanderthals interbred with ancestors of today's Europeans when modern humans began spreading out of Africa — 1.5 to 2.1 percent of the DNA of anyone living outside Africa today is Neanderthal in origin. [Image Gallery: Our Closest Human Ancestor] In 1993, scientists found an extraordinarily intact skeleton of an ancient human amidst the stalactites and stalagmites of the limestone cave of Lamalunga, near Altamura in southern Italy — a discovery they said had the potential to reveal new clues about Neanderthals. "The Altamura man represents the most complete skeleton of a single nonmodern human ever found," study co-author Fabio Di Vincenzo, a paleoanthropologist at Sapienza University of Rome, told Live Science. "Almost all the bony elements are preserved and undamaged."  The Altamura skeleton bears a number of Neanderthal traits, particularly in the face and the back of the skull. However, it also possesses features that usually aren't seen in Neanderthals — for instance, its brow ridges were even more massive than those of Neanderthals.These differences made it difficult to tell which human lineage the Altamura man might have belonged to. Moreover, the Altamura skeleton remains partially embedded in rock, making it difficult to analyze. Now, new research shows that DNA from a piece of the skeleton's right shoulder blade suggests the Altamura fossil was a Neanderthal. The shape of this piece of bone also looks Neanderthal, the researchers said. In addition, the scientists dated the skeleton to about 130,000 to 170,000 years old. This makes it the oldest Neanderthal from which DNA has ever been extracted. (These bones are not the oldest known Neanderthal fossils — the oldest ones ever found are about 200,000 years old. This isn't the oldest DNA ever extracted from a human, either; that accolade goes to 400,000-year-old DNA collected from relatives of Neanderthals.) The bone is so old that its DNA is too degraded for the researchers to sequence the fossil's genome — at least with current technology. However, they noted that next-generation DNA-sequencing technologies might be capable of such a task, which "could provide important results on the Neanderthal genome," study co-author David Caramelli, a molecular anthropologist at the University of Florence in Italy, told Live Science.  Whereas previous fragmentary fossils of different Neanderthals provided a partial picture of what life was like for Neanderthals, the Altamura skeleton could help paint a more complete portrait of a Neanderthal — for instance, it could reveal more details about Neanderthals' genetics, anatomy, ecology and lifestyle, the researchers said. "We have a nearly complete human fossil skeleton to describe and study in detail. It is a dream," Di Vincenzo said. "His morphology offers a rare glimpse on the earliest phase of the evolutionary history of Neanderthals and on one of the most crucial events in human evolution. He can help us better understand when — and, in particular, how — Neanderthals evolved." “Despite the fact that this specimen represents one of the most extraordinary hominin specimens ever found in Europe, for the last two decades our knowledge of it has been based purely on the documented on-site observations,” the authors wrote in the abstract. “Recently, the retrieval from the cave of a fragment of bone (part of the right scapula) allowed the first dating of the individual, the quantitative analysis of a diagnostic morphological feature, and a preliminary paleogenetic characterization of this hominin skeleton from Altamura.” Source: Lari M, Di Vincenzo F, Borsato A, Ghirotto S, Micheli M, Balsamo C. The Neanderthal in the karst: First dating, morphometric, and paleogenetic data on the fossil skeleton from Altamura (Italy). Journal of Human Evolution. 2015. |
|
|
|
Post by Admin on Apr 17, 2015 14:34:19 GMT
 Analyses of morphological traits, Neandertal ancient DNA and modern DNA (e.g., refs. 7–9), appear to support a recent African origin of all humankind. However, it has been argued that patterns of genetic diversity are not incompatible with a multiregional model (5, 10–12). For instance, Nordborg (10) showed that the differences between Neandertal's and modern mitochondrial sequences are sufficient to rule out random mating between them, but not more complicated models of interbreeding. Those results reflect the existing uncertainties on the European demographic history of the last 30,000 years. Clearly, genetic typing of the earliest a.m.h. in Europe, sometimes referred to as Cro-Magnons or Cro-Magnoid from the site in France where they were first discovered, is a crucial step for solving this question (13–15), because that would allow a genetic comparison between individuals who lived at a much shorter (ideally, zero) time distance. The Out-of Africa model, in fact, predicts genetic discontinuity between Neandertals and early a.m.h. (the former being a separate lineage replaced by the latter) and genetic continuity along the a.m.h. lineages from the Upper Palaeolithic until the present. The multiregional models, on the contrary, predict at least some level of genetic continuity from the archaic Neandertal forms to the almost contemporary Cro-Magnon forms up to today's Europeans.  Fig. 1. MDS of HVRI sequences of 60 modern Europeans (filled squares), 20 modern non-Europeans (filled circles), 4 Neandertals (open diamonds), the Australian Lake Mungo 3 (open circle), and the two early a.m.h. typed in this study (open squares). European and non-European sequences in this figure were selected to represent the most divergent lineages observed in modern individuals. Note that the axes have different scales. The stress value for this analysis was 0.128. Specific mtDNA sites outside HVRI were also analyzed (by amplification, cloning, and sequencing of the surrounding region) to classify more precisely the ancient sequences within the phylogenetic network of present-time mtDNAs (35, 36). Paglicci-25 has the following motifs: +7,025 AluI, 00073A, 11719G, and 12308A. Therefore, this sequence belongs to either haplogroups HV or pre-HV, two haplogroups rare in general but with a comparatively high frequencies among today's Near-Easterners (35). Paglicci-12 shows the motifs 00073G, 10873C, 10238T, and AACC between nucleotide positions 10397 and 10400, which allows the classification of this sequence into the macrohaplogroupN,containing haplogroups W, X, I, N1a, N1b, N1c, and N*. Following the definition given in ref. 36, the presence of a single mutation in 16,223 within HRVI suggests a classification of Paglicci-12 into the haplogroup N*, which is observed today in several samples from the Near East and, at lower frequencies, in the Caucasus (35). It is difficult to say whether the apparent evolutionary relationship between Paglicci-25 and Paglicci-12 and those populations is more than a coincidence. Indeed, the haplogroups to which the Cro-Magnon type sequences appear to belong are rare among modern samples, and therefore their frequencies are poorly estimated. However, genetic affinities between the first anatomically modern Europeans and current populations of the Near East make sense in the light of the likely routes of Upper Paleolithic human expansions in Europe, as documented in the archaeological record (37). A pattern of genetic distances through time emerges clearly when four additional HVRI sequences from prehistoric anatomically modern Europeans dated between 5,500 and 14,000 years ago (38, 39) are considered (Fig. 2). Going back in time from the present to 25,000 years ago, prehistoric Europeans show an approximately constant number of differences in comparison with today's Europeans, very similar indeed to the number of differences between two randomly chosen modern sequences. Conversely, an abrupt increase of genetic distance from today's Europeans is observed for the Neandertals, even though the most recent of them is separated from the older Paglicci sample by only a few hundreds of generations. Therefore, these results suggest a pattern of genetic continuity in the modern humans' genealogy from the Upper Palaeolithic period to the present, but a clear discontinuity with respect to Neandertals of similar ages.  Fig. 2. Average genetic distance between ancient and modern samples (2,566 sequences of modern Europeans; y axis), as a function of the samples' age (x axis, in thousands of years). Vertical lines represent two standard deviations above and below the mean. Squares, a.m.h. Diamonds, Neandertals. The Paglicci samples typed in this study are indicated by open squares. The point at 0 years indicates the average pairwise difference between present-day samples. Even the most stringent available criteria for validating ancient human DNA sequences do not allow one to prove that the sequences determined are authentic. Only if a sequence is radically different from modern ones, as is the case for Neandertals, can one be relatively sure that no contamination has affected the results. Therefore, a certain degree of prudence is necessary before drawing any conclusions from this study. Still, none of the biochemical tests we carried out suggests that different sequences (namely the endogenous one plus some contaminating sequences) were amplified from the 23,000- and 25,000-year-old specimens that we used. In addition, the amino acid racemization test strongly suggests that reasonably well preserved DNA should be present in those specimens. Because DNA from all four Cro-Magnon type bone fragments could be amplified and sequenced only by using primers specific for modern humans, and not for Neandertals, there is little doubt that the mtDNAs of early a.m.h. and of cronologically close Neandertals were, at least, very different. Under the multiregional model of human origins, Neandertals and modern humans are just one population observed at different times (1, 2). Therefore, when comparing sequences sampled along the single human–Neandertal genealogy, one should not observe any major discontinuity. More recent versions of the multiregional model (3–5) suggest that Neandertals and early a.m.h. were regional populations of the same evolving species connected by gene flow, and both archaic and modern forms contributed, possibly in different proportions, to the present day human gene pool. Under this updated multiregional model, the absence of Neandertal mtDNA lineages in living humans is regarded as a consequence of a random drift or a selection process of lineage extinction since the disappearance of Neandertals. In this case, unless the extinction of Neandertal lineages was almost instantaneous, the probability of finding such lineages in early a.m.h. should not be too low. All these expectations are inconsistent with the data and the analysis here presented. Two a.m.h. dated between 23,000 and 25,000 years ago appear to have HVRI sequences fully compatible with the variation observed both in contemporary and in ancient samples of a.m.h., and certainly they do not show any special relationships with the almost contemporary Neandertals. These results are at odds with the view whereby Neandertals were genetically related with the anatomically modern ancestors of current Europeans or contributed to the present day human gene pool. Although only six HVRI sequences of ancient a.m.h and four sequences of Neandertals are available to date, the sharp differentiation among them represents a problem for any model regarding the transition from archaic to modern humans as a process taking place within a single evolving human lineage. Caramelli, David, et al. " Evidence for a genetic discontinuity between Neandertals and 24,000-year-old anatomically modern Europeans." Proceedings of the National Academy of Sciences 100.11 (2003): 6593-6597. |
|
|
|
Post by Admin on Apr 21, 2015 14:19:06 GMT
 The Lamalunga cave opens in the limestone of the Murgia plateau at an elevation of 508 m a.s.l., near the town of Altamura (Puglia, Italy; Agostini, 2011). It constitutes the upper part of a larger karstic complex where stalactites, stalagmites, and flowstones occur together with “coralloid” formations, which mostly represent the last phase of calcite precipitation caused by spray/aerosol phenomena. This complex consists mainly of a sub-horizontal gallery that had developed at a shallow depth from the surface, intercepted by pits that had originally opened to the surface but which have subsequently been clogged by detritus. In this context, the discovery of a virtually complete fossilized hominin skeleton in an excellent state of preservation gives rise to interesting taphonomic considerations. Particularly, faunal remains found in some of the galleries are often isolated bony elements accumulated in depressed areas of the cave, suggesting that they were transported and dispersed by water. This was not the case with the human skeleton, given that it is largely represented and concentrated in a small area. Thus, we may hypothesize that, after death and decomposition of the body, the skeleton collapsed where it has been found. Thus far, no lithic tools have been found in the cave.  Figure 1. A) Position of Altamura within the Italian peninsula; B) hominin bones and calcite formations around the cranium (part of the mandible and right femur are visible); C) general topography of the northern part of the Lamalunga karstic system; note on the left the accumulation of detritus that represents the infilling of the probable main original access point from the external surface; and D) distribution of the main bones of the skeleton at the end of the so-called “ramo dell'uomo” (compare SOM Fig. 1). Drawing and data of Fig. 1D are from Vacca and Pesce Delfino (2004). Even though the skeleton is partly incorporated into calcite concretions and is covered by coralloid formations, most of the bones are visible (see Fig. 1; see also Supporting Online Material [SOM] Fig. 1), including the cranium (upside down), the mandible, and several postcranial elements. From the photographs available and direct observations made in situ by one of us (GM), the skeleton appears to exhibit a mixture of archaic and derived features, which fit the range of variation typical of European hominins of the late Middle/early Late Pleistocene (Manzi et al., 2011). In fact, even though a number of Neanderthal traits can be seen—particularly in the face and in the occipital bone—there are features that distinguish this specimen from the more typical morphology of Homo neanderthalensis, such as the shape of the brow ridges, the relative dimension of the mastoids, and the general architecture of the cranial vault. U/Th dating and petrography A previous series of 25 U/Th dating was carried out by alpha spectrometry on a series of stalactites, flowstones, and coralloids (Fig. 2) by Branca and Voltaggio (2011). These revealed an ancient phase of speleothem formation, dating to between 189 ± 29 and 172 ± 15 ka, and a second phase, indicated by some of the flowstones, between 45.9 ± 1.7 and 34.4 ± 1.5 ka, while the ages of the coralloids (13 analyses in all) were distributed continuously between 43.3 ± 1.6 and 29.1 ± 1.0 ka and between 17.1 ± 0.7 ka and 13.4 ± 0.5 ka. Outside of these age groups, two additional single dates were recorded: 133 ± 9 ka for a stalactite and 98.7 ± 4.4 ka for a flowstone.  Figure 2. Synthesis of the Alpha spectrometry ages on speleothem from the Lamalunga cave and the new MC-ICP-MS analyses versus their U content; the error bars of the MC-ICP-MS analyses (2σ) are always smaller than the plotted symbols. (* Data from Branca and Voltaggio, 2011). The microstratigraphy of the newly collected calcite samples ABS3 and ABS5 highlights four growth phases separated by three discontinuities (Fig. 3). The oldest growth phase, directly coating the hominin bone in ABS3 and the broken stalagmite in ABS5, is characterized by micrite laminae grading into microsparite and columnar calcite. This small similar pattern suggests that the two layers could be correlated and have a similar age. The other layers consist of elongated open and compact columnar calcite (cf. Frisia et al., 2000 and Frisia and Borsato, 2010). On the basis of the identical microstratigraphy in the two samples, a similar micro-sampling strategy was performed in order to double check the age of the calcite overgrowth on the hominin bone. Moreover, since the overgrowth on ABS5 is considerably thicker than that on ABS3, this approach permitted avoidance of possible contamination from adjacent layers. We performed six MC-ICP-MS analyses on the three calcite overgrowths over the hominin bones and four analyses on the corresponding growth phases identified on the overgrowth on stalagmite ABS5 (Fig. 3). The results (Table 1) revealed four growth episodes dated to 7.04 ± 0.72–7.6 ± 0.04 ka, 36.9 ± 0.17–38.1 ± 0.61 ka, 64.6 ± 0.33 ka, and 121.9 ± 2.22–130.1 ± 1.9 ka, respectively corresponding to the warm Marine Isotope Stages (MIS) 1.0, 3.1, 5.1, and 5.5 (cf. Martinson et al., 1987) and matching coeval speleothem growth phases in Mediterranean caves (Bar-Matthews et al., 2003 and Badertscher et al., 2011). The correlation between speleothem growth phases in Altamura cave and warm MIS gives rise to an important consideration: the warm Marine Isotope Stage 7.1, between ca. 185 and 200 ka (Martinson et al., 1987), was not recorded in the coralloid overgrowths over the hominin bones, although it was recorded in two other stalactites from the same cave chamber (growth phase between 189 ± 29 and 172 ± 15 ka; cf. Fig. 2), as well as in speleothems from other Mediterranean caves (Bar-Matthews et al., 2003 and Badertscher et al., 2011). Given the fact that all the other growth phases are represented in overgrowth ABS3, this suggests that the hominin bones could be more recent than 172 ± 15 ka.  Figure 3. Selected calcite crusts and coralloid overgrowths with the calculated U/Th ages: A) thin calcite crust coating the underside of a long bone (fibula; ABS2); B) polished slab of mm-thick coralloid overgrowth covering the termination of a short bone (ABS3); and C) thin section of coralloid mm-thick overgrowth covering a naturally broken stalagmite (ABS5). The dotted, dashed, and dotted-dashed red lines on ABS3 and ABS5 visualize similar discontinuities that define the different growth phases identified by the U/Th ages. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) Morphometric analysis Metrical variables show that the scapulo-humeral joint of the Altamura skeleton is similar to those of European samples from the Middle and Upper Pleistocene with respect to the diameter and depth of the SGF (as reported in the SOM Fig. 5). Geometric morphometrics may be used to better characterize the shape of the SGF, which has diagnostic significance (Di Vincenzo et al., 2012). When 2D landmark data are analyzed by Principal Component Analysis (PCA; Fig. 4B), the distribution of the samples reveals a signal of taxonomic and phylogenetic significance. Notably, the linear regression of the centroid size values on the 1st PC (r = −0.23; p = 0.057), as well as the multivariate regression of the centroid sizes on all the PC scores (p = 0.522) are not significant; thus, our results are not significantly influenced by allometry.  Figure 4. A) The articular portion of the right scapula from Altamura, viewed from the glenoid fossa (SGF), with landmarks (darker/green points in the online version of the paper) and semilandmarks (lighter/yellow points); B) Variance along PC1 and PC2 of the whole sample; OTUs are bounded by lines, while labels of the specimens are as in SOM Table 1; deformation grids represent extreme shape variations. C) Neighbour Joining of the phenetic relationships between the averaged OTUs and the specimen from Altamura. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) Paleogenetic analysis Despite several PCR attempts, it was only possible to obtain positive results from one of the shortest amplicons (82 bp; see SOM Fig. 6). All sequence results, obtained in two different laboratories, were concordant and presented a typical Neanderthal mtDNA haplotype (Condemi et al., 2013; A16230G, G16244A, C16256A, A16258G; Fig. 5). The consensus sequence was deposited in GenBank under the accesion number KJ888153. Despite its short length, the mtDNA fragment from Altamura falls within the known range of Neanderthal diversity, being clearly distinct from both modern humans and the Denisova sequences. The sample from Atapuerca Sima de los Huesos (Meyer et al., 2013) was excluded from the subsequent analysis because for the fragment considered here it is poorly covered and misses informative sites. The sequence from Altamura clusters together with other Neanderthals from Western Europe in the tree reported in Figure 6. This picture seems to confirm the existence of some degree of population structuring among Neanderthals, already present around 150 ka. Unfortunately, the genetic data used in the analysis are too scant to make robust inferences regarding patterns of temporal and/or geographic variation in Neanderthal genetic diversity.  Figure 6. Neighbor joining tree showing the phylogenetic relationships among Neanderthals on the basis of the present analysis. The name of the samples are reported next to their accession number (if present); Denisova is used as an outgroup. Conclusions Overall, the results of our morphometric and the paleogenetic analyses concur in indicating that the skeleton from Altamura belongs to a Neanderthal. In addition, using U/Th dating we were able to provide the first range of dates for the specimen, between 130 ± 2 ka and 172 ± 15 ka. Nevertheless, some features exhibited by the skeleton and observed in situ (on the cranium, in particular, as summarized in the Introduction) differ from the morphology known among the typical representatives of Homo neanderthalensis, while they appear consistent with the pre-Würmian age we obtained. Metrical variables show that the scapula-humeral joint is closer to the morphotype usually referred to the so-called “early Neanderthals,” including specimens such as those from Saccopastore (e.g., Bruner and Manzi, 2006), Krapina (e.g., Monge et al., 2008), or Apidima ( Harvati et al., 2011). In addition, geometric morphometric analysis of the SGF from Altamura suggests some peculiarities of this small piece of bone, while (consistent with the mtDNA data) the same analysis strengthens the notion that the Neanderthal morphology was essentially present in the late Middle Pleistocene. Finally, it is of great interest that mtDNA was sufficiently preserved to permit paleogenetic analysis. The results of the explorative approach used here have shown that the sample contained endogenous DNA (although highly fragmented) with a typical Neanderthal haplotype; moreover, there was no evidence of modern human contamination in the bone fragment, at least not at the mtDNA level. For these reasons, the Altamura skeleton should be considered a good candidate for more innovative genomic analyses, like capture approaches or ultra-deep shotgun sequencing, especially when we consider that Altamura represents the most ancient Neanderthal from which endogenous DNA has been retrieved so far. doi:10.1016/j.jhevol.2015.02.007 |
|
|
|
Post by Admin on Apr 25, 2015 5:55:00 GMT
Tens of thousands of years ago, the rise of humanity as we know it began. Modern humans spread from Africa to Europe and Asia, which were already populated by older Neanderthal cultures. The two groups interbred, adding some Neanderthal DNA to our present-day genes. The relatively sophisticated humans flourished, while their Neanderthal counterparts disappeared, going extinct in Europe somewhere between 39,000 to 41,000 years ago. But the timeline, in many places, is still foggy. By identifying a pair of ancient teeth as conclusively human, a team of scientists think they've managed to clear things up a little — and helped settle a debate about how different human and Neanderthal cultures might have been. "From 45,000 up to 40,000 [years], we have several cultures that suddenly appeared in Europe," says University of Bologna researcher Stefano Benazzi, co-author of a paper that appears today in Science. One of those cultures was the Protoaurignacian, which developed in southern Europe around 42,000 years ago. The Aurignacian culture that followed marked a turning point in modern humanity: among other things, it gave us the world's earliest known musical instruments, the earliest known wall art, and potentially the first representation of a human figure. Protoaurignacians themselves are known for making personal ornaments, small stone blades, and other things that are "quite typical of modern humans." But while their artifacts have been left behind, it's hard to conclusively prove that the Protoaurignacians actually were human. /cdn0.vox-cdn.com/uploads/chorus_asset/file/3635750/benazzi5HR.0.jpg) In fact, only three remains have ever been recovered from known Protoaurignacian sites: a fragment of fetal bone from France and two teeth, found in different parts of northern Italy in 1976 and 1992. It was widely expected that they came from humans, Benazzi says, but they couldn't rule out the possibility that Neanderthals had managed to develop a strikingly modern culture. "It would mean that Neanderthals had exactly the same skill as modern humans," says Benazzi, and could weaken the theory that humans had survived by outcompeting them. In order to test this, Benazzi and his team examined the two tooth samples. First, they checked the enamel of one against a range of newer human teeth. Neanderthal teeth have comparatively thin enamel layers, and even heavily worn down, the Protoaurignacian tooth was closer to human measurements. For the second tooth, they managed to extract mitochondrial DNA, which sits outside a cell's nucleus and is passed down only through the mother. Checked against the DNA of 54 present-day humans, 10 ancient modern humans, and 10 Neanderthals, along with another extinct species of human and a chimpanzee, it fell squarely within the modern human range.  Unfortunately, because mitochondrial DNA comes only from the mother, the team can't get a full picture of the genetics. "From enamel thickness and the mitochondrial DNA, we cannot exclude interbreeding," says Benazzi. The mother of whomever owned the second tooth was human, he says, "but if the father of this child was a Neanderthal, we don't know." His team hopes to later extract nuclear DNA, which contains both parents' genes, but that's a more difficult proposition. Even if it turns up conclusively human, the study is limited by the fact that there are only two samples. Considering that it's taken nearly 40 years to find even these two, they can't count on having another one turn up. Based on the estimated age of the two teeth, these humans lived in southern Europe at least 41,000 years ago, around the time Neanderthals went extinct. Though it's only speculative, Benazzi believes this supports the idea that modern humans contributed to their demise, whether through direct measures or some kind of indirect competition. No matter what, he thinks they've demonstrated that the Protoaurignacian culture can be traced back to modern humans — not, at least exclusively, highly skilled Neanderthals. Science DOI: 10.1126/science.aaa2773 |
|
|
|
Post by Admin on May 3, 2015 4:57:45 GMT
 Analyses of Neanderthal genomes indicate that when anatomically modern humans ventured out of Africa around 50,000 years ago, they met and mated with Neanderthals, probably in regions of the Eastern Mediterranean. We know that Neanderthals inhabited regions of Eurasia during the recent ice ages for a period of over 200,000 years and finally became extinct around 40,000 to 35,000 years ago. Today, when we examine the genomes of Europeans and Asians, we find that about 2% of their genomes consist of Neanderthal fragments. Africans either do not have or have very small percentages of Neanderthal DNA, probably due to limited interbreeding between Eurasian peoples and Africans in more recent times. In general, the fraction of Neanderthal DNA in Eurasians is very small, indicating that modern humans interbred with Neanderthals only over a limited geographic area and period of time, perhaps in the zone where we first met. Additionally, it is possible there was rather strong avoidance to mating, perhaps based on different appearances or social behaviors. Moreover, it is quite possible that hybrids between Neanderthals and modern humans suffered reduced fertility. Though the fraction is small, genomes are vast. Thus, even 2% Neanderthal DNA in Eurasian genomes totals nearly 60 million bases of DNA – the A, C, G and Ts. So, what are the functional effects of this Neanderthal DNA on our biology? Was the DNA we inherited from our ancient Neanderthal parents harmful to us or was it beneficial to us?  Regions of Neanderthal ancestry on chromosome 12 in the Ust’-Ishim individual and fifteen present-day non-Africans. In recent research by Svante Pääbo and coworkers, this question was approached by analyzing how the 2% Neanderthal DNA is distributed among the over three billion bases in our genomes. Through previous studies of our genome, we know that only a very small fraction is functional, meaning it codes or regulates proteins in our body. Instead, vast stretches of our genome seem to be non-functional and have no biological effects. If Neanderthal DNA was in general not harmful to us, we would expect nearly equal percentages in both functional and non-functional parts of our genome. Alternatively, if Neanderthal DNA is more detrimental, we would expect to see less of it in functional regions and more of it in non-functional regions. Indeed, this is exactly what is found. Pääbo and coworkers discovered significantly less Neanderthal DNA in functional parts of our genome and more of it in non-functional parts. Moreover, they found that the greatest “desert” of Neanderthal DNA in human genomes is on the X chromosome where they found five times less Neanderthal DNA compared to other chromosomes. How did the Neanderthal segments become distributed in such a lopsided way within our genome? One way is through the action of negative selection, that process of natural selection that weeds out harmful DNA. Over the 2,000 generations since humans interbred with Neanderthals, negative selection seems to have been hard at work removing Neanderthal DNA from the functional parts of our genome though leaving it in regions where it has no biological effect. What could account for the dramatic reduction of Neanderthal DNA on human X chromosomes? The pattern could be explained if male hybrid offspring between mating’s of modern humans and Neanderthals had reduced fertility. Genes located on male hybrid X chromosomes may not have been compatible with the set of chromosomes inherited from the “other” parent. It is well known that hybrids of the heterogametic sex (the male sex in most mammals, having X and Y chromosomes) suffer reduced fertility much more than female hybrids do. The thinking is that because female hybrids have two X chromosomes, any one of these can cover up for harmful effects the other X chromosome might cause, whereas males with only one X chromosome are left “exposed.” Consistent with the idea that male hybrids had low fertility is the finding that human genes expressed in testes contain 20 to 100 times less Neanderthal DNA compared to genes expressed in other organs such as kidney, liver, heart, or brain. 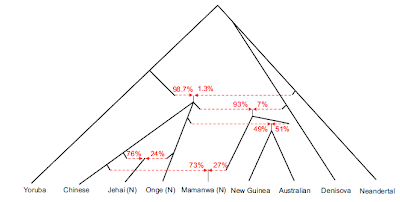 So, was any of the DNA we inherited from Neanderthals good for us? Modern humans spread relatively rapidly into territories of Eurasia, regions that Neanderthals had already inhabited for over 200,000 years. Therefore, it’s probably true that Neanderthals had already become sufficiently adapted to survive in Eurasian environments and that some of their genes would be beneficial to modern humans. Upon interbreeding with Neanderthals, these favorable genes could have spread within modern human populations by a second type of natural selection, called positive natural selection. Although work is ongoing, and we undoubtedly have much to learn, when we analyze large numbers of genomes from Europeans and Asians (determined by the ongoing 1000 Genomes Project), certain regions inherited from Neanderthals bear signs that they are biologically functional in human individuals, and in some cases even became more common in Eurasian populations suggesting they were beneficial. The list of potentially beneficial Neanderthal genes fall into several categories: genes coding for aspects of skin and hair, genes involved in galactose metabolism (possibly related to a newborn’s ability to digest mother’s milk), genes related to long-term depression (possibly related to cerebellar learning), genes involved in immune defense, and genes involved in lipid-digestion. That modern humans inherited immune defense genes and lipid-digestion genes from Neanderthals does not seem too surprising as humans would have benefited greatly from any genes that helped them fight new pathogens and or digest new foods they encountered as they colonized Europe and Asia. Skin and hair genes relate to pigmentation, skin-cell differentiation, and the skin-protein keratin. With regard to keratin genes, it appears that different sets of keratin genes from Neanderthals were greatly favored in different populations. We are not sure of the reasons, but Europeans seemed to have favored Neanderthal-derived genes coding for keratin filaments in skin, whereas East Asians favored genes linked to keratin filaments in hair. There is much we have to learn about the Neanderthal DNA in us. Through investigating some its harmful effects, we might get a better handle on why Neanderthals ultimately vanished. On the other hand, it is heartening to know that at least some Neanderthal DNA in our genomes seems to have helped us along our evolutionary journey. |
|













/cdn0.vox-cdn.com/uploads/chorus_asset/file/3635750/benazzi5HR.0.jpg)



