Post by Admin on Sept 24, 2021 0:34:33 GMT
Results
Data
Here we present a new genomic dataset sampled from the Leeward Society Islands, eastern Polynesia. We report high-resolution autosomal genotyping data from 30 individuals, complemented by genotyping and/or re-sequencing of uniparental loci (mtDNA and Y) from 81 individuals, including seven Y chromosomes re-sequenced by a target-capture method. In addition, we present new uniparental data from 49 Maori individuals sampled in New Zealand (Supplementary Tables S1–S3). The dataset is analyzed together with publicly available data from Island Southeast Asia, Melanesia and Polynesia (Supplementary Tables S1, S4 and S5). For detailed information about samples used in the present study, please refer to the Materials and Methods section.
Autosomal analysis
The first two PCs of the principal components analysis (PCA, Fig. 1c) account for 38% of the variation in the studied dataset. The close overlap between eastern Polynesians and Samoans on the PC1 axis suggests similar amounts of genetic ancestry shared with New Guinea and northern Melanesia. The model-based analysis of autosomal SNPs using ADMIXTURE35 shows that, at K = 4, 70–80% of the Leeward Society Islander genomes can be characterized by the component typical of ISEA/East Asia (Fig. 2a); the remaining 20–30% of their genetic ancestry is best represented by Papuan speakers from New Guinea (light purple). From K = 5, Polynesians take their own ancestry (green), which, like their deflection on the PCA plot, is most likely due to genetic drift or, alternatively, cryptic relatedness or extreme inbreeding in studied populations. However, the latter is unlikely due to the lack of close relatives (up to third-degree, inclusive) in four Polynesian groups, and normal range of inbreeding coefficients when comparing to other human populations (F IS , Supplementary Table S6).
Figure 2
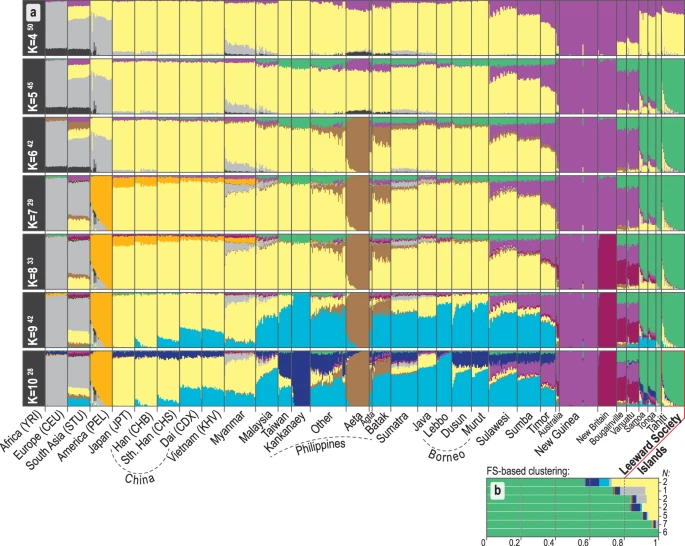
Ancestral genomic components in study populations estimated using ADMIXTURE. Details of the populations are provided in Supplementary Table S1B. The colors used have been selected to be equivalent to those used in Fig. 1. Only runs from K = 4 to 10 are shown, complete results (K = 2 to 15) are given in Supplementary Fig. S1. (a) For every value of K, the modal solution with the highest number (superscript) of ADMIXTURE35 runs is shown; individual ancestry proportions were averaged across all runs and the average cross-validation statistics were calculated across all runs from the same mode (Supplementary Fig. S2). The minimum cross-validation score is observed at K = 11 but no further components appear in the profiles of Polynesians after K = 10. Populations from the Philippines can be generally divided into Negritos (Aeta, Agta and Batak), Kankanaey of northwestern Luzon, and all others representing an amalgamation of groups from Luzon, Palawan and Visayas (see Fig. 1 and Supplementary Table S1B). (b) The average of K = 10 ADMIXTURE profiles for groups of Leeward Society individuals clustered by fineSTRUCTURE37 (Supplementary Fig. S6), indicating the heterogeneous distribution of East Asian and European ancestry among the Leeward Society Islanders.
The lowest cross-validation (CV) score of ADMIXTURE is observed at K = 11, but no additional ancestries appear in Polynesians after K = 10, which has the second lowest CV score (Fig. 2a, Supplementary Figs S1 and S2). At K = 10, a dark blue component appears that is almost fixed in the Kankanaey of northwestern Luzon. The distinctive and uniform profiles of additional ISEA, Melanesian, and East Asian ancestries in two (Tonga and Samoa) out of four, otherwise very closely related, Polynesian groups hint that these may be the result of an old admixture process, rather than genetic drift, extreme bottlenecks or algorithmic artifacts. In contrast, the noticeably uneven distribution of the East Asian (yellow) and western European (grey) ancestry components within the profiles of the Leeward Society individuals (Fig. 2b) is consistent with recent historical admixture events (see haplotype-based admixture analysis below).
The outgroup f336 allele-sharing plot shows the length of a phylogenetic branch shared between two study populations and African Yoruba. For the Leeward Society Isles (Supplementary Fig. S3, Supplementary Table S7), the f3 allele-sharing results are consistent with a most recent evolutionary history shared with Samoa, Tahiti, and Tonga. It also suggests that the Kankanaey of the Philippines and Taiwanese aborigines are the next closest populations to all four Polynesian groups. These results remain robust to the different SNP subsets or population clustering schemes used in the present study (Supplementary Figs S3, S4, Supplementary Table S7). In contrast, the f3 admixture plots (Supplementary Fig. S5, Supplementary Table S7), which detect the presence of admixture in a study population from two reference groups, display different results for western and eastern Polynesia. These differences could be explained by a reduced effective population size for eastern Polynesians, caused by bottlenecks during the initial settlement process, or because Tonga and Samoa have experienced additional admixture since they last shared a common ancestral gene pool with Tahiti and the Leeward Society Isles.
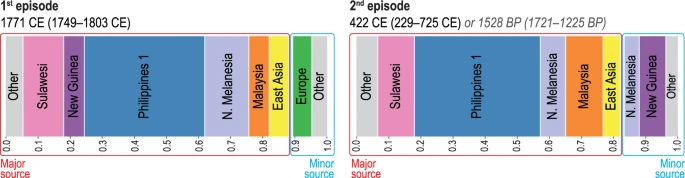
The unsupervised fineSTRUCTURE (FS) analysis of haplotypes37 placed individuals into genetic clusters that include: Philippine groups from lowland Luzon, Palawan, and Visayas (‘Philippines 1’), Malaysia, Sulawesi, East Asia, northern Melanesia (Bougainville), New Guinea, and western Europe (Supplementary Fig. S6). The GLOBETROTTER (GT) analysis38 produced strong statistical support for two separate episodes of admixture involving the ancestors of the Leeward Society Islanders (Fig. 3, Supplementary Table S8). The first represents an average contribution of ~6% western European ancestry, which is dated to 1749–1803 CE. This is consistent with documented contact during Cook’s three voyages of exploration1, which took place 1768-71, 1772-75 and 1776-80. The second episode is estimated to have occurred in an interval from ~1,200 to 1,700 BP (229–725 CE), and is composed of a minor component (~17%), comprising mainly northern Melanesian and New Guinea sources, and a major one (~83%), in which the largest contributions are attributable to the ‘Philippines 1’, Sulawesi, and Malaysian clusters. The chronology indicates that this episode occurred prior to the earliest widely accepted radiometric dates for the permanent settlement of eastern Polynesia, which centre on ~950 BP and come from archaeological sites on Rai’atea in the Leeward Society Isles26,27. In addition, the presence of northern Melanesian ancestry in the minor component of the second (older) episode of admixture (~8% of the genome) reflects some genetic contact with this region for the ancestors of the Leeward Society Islanders prior to 1,200–1,700 BP.
Data
Here we present a new genomic dataset sampled from the Leeward Society Islands, eastern Polynesia. We report high-resolution autosomal genotyping data from 30 individuals, complemented by genotyping and/or re-sequencing of uniparental loci (mtDNA and Y) from 81 individuals, including seven Y chromosomes re-sequenced by a target-capture method. In addition, we present new uniparental data from 49 Maori individuals sampled in New Zealand (Supplementary Tables S1–S3). The dataset is analyzed together with publicly available data from Island Southeast Asia, Melanesia and Polynesia (Supplementary Tables S1, S4 and S5). For detailed information about samples used in the present study, please refer to the Materials and Methods section.
Autosomal analysis
The first two PCs of the principal components analysis (PCA, Fig. 1c) account for 38% of the variation in the studied dataset. The close overlap between eastern Polynesians and Samoans on the PC1 axis suggests similar amounts of genetic ancestry shared with New Guinea and northern Melanesia. The model-based analysis of autosomal SNPs using ADMIXTURE35 shows that, at K = 4, 70–80% of the Leeward Society Islander genomes can be characterized by the component typical of ISEA/East Asia (Fig. 2a); the remaining 20–30% of their genetic ancestry is best represented by Papuan speakers from New Guinea (light purple). From K = 5, Polynesians take their own ancestry (green), which, like their deflection on the PCA plot, is most likely due to genetic drift or, alternatively, cryptic relatedness or extreme inbreeding in studied populations. However, the latter is unlikely due to the lack of close relatives (up to third-degree, inclusive) in four Polynesian groups, and normal range of inbreeding coefficients when comparing to other human populations (F IS , Supplementary Table S6).
Figure 2

Ancestral genomic components in study populations estimated using ADMIXTURE. Details of the populations are provided in Supplementary Table S1B. The colors used have been selected to be equivalent to those used in Fig. 1. Only runs from K = 4 to 10 are shown, complete results (K = 2 to 15) are given in Supplementary Fig. S1. (a) For every value of K, the modal solution with the highest number (superscript) of ADMIXTURE35 runs is shown; individual ancestry proportions were averaged across all runs and the average cross-validation statistics were calculated across all runs from the same mode (Supplementary Fig. S2). The minimum cross-validation score is observed at K = 11 but no further components appear in the profiles of Polynesians after K = 10. Populations from the Philippines can be generally divided into Negritos (Aeta, Agta and Batak), Kankanaey of northwestern Luzon, and all others representing an amalgamation of groups from Luzon, Palawan and Visayas (see Fig. 1 and Supplementary Table S1B). (b) The average of K = 10 ADMIXTURE profiles for groups of Leeward Society individuals clustered by fineSTRUCTURE37 (Supplementary Fig. S6), indicating the heterogeneous distribution of East Asian and European ancestry among the Leeward Society Islanders.
The lowest cross-validation (CV) score of ADMIXTURE is observed at K = 11, but no additional ancestries appear in Polynesians after K = 10, which has the second lowest CV score (Fig. 2a, Supplementary Figs S1 and S2). At K = 10, a dark blue component appears that is almost fixed in the Kankanaey of northwestern Luzon. The distinctive and uniform profiles of additional ISEA, Melanesian, and East Asian ancestries in two (Tonga and Samoa) out of four, otherwise very closely related, Polynesian groups hint that these may be the result of an old admixture process, rather than genetic drift, extreme bottlenecks or algorithmic artifacts. In contrast, the noticeably uneven distribution of the East Asian (yellow) and western European (grey) ancestry components within the profiles of the Leeward Society individuals (Fig. 2b) is consistent with recent historical admixture events (see haplotype-based admixture analysis below).
The outgroup f336 allele-sharing plot shows the length of a phylogenetic branch shared between two study populations and African Yoruba. For the Leeward Society Isles (Supplementary Fig. S3, Supplementary Table S7), the f3 allele-sharing results are consistent with a most recent evolutionary history shared with Samoa, Tahiti, and Tonga. It also suggests that the Kankanaey of the Philippines and Taiwanese aborigines are the next closest populations to all four Polynesian groups. These results remain robust to the different SNP subsets or population clustering schemes used in the present study (Supplementary Figs S3, S4, Supplementary Table S7). In contrast, the f3 admixture plots (Supplementary Fig. S5, Supplementary Table S7), which detect the presence of admixture in a study population from two reference groups, display different results for western and eastern Polynesia. These differences could be explained by a reduced effective population size for eastern Polynesians, caused by bottlenecks during the initial settlement process, or because Tonga and Samoa have experienced additional admixture since they last shared a common ancestral gene pool with Tahiti and the Leeward Society Isles.
The unsupervised fineSTRUCTURE (FS) analysis of haplotypes37 placed individuals into genetic clusters that include: Philippine groups from lowland Luzon, Palawan, and Visayas (‘Philippines 1’), Malaysia, Sulawesi, East Asia, northern Melanesia (Bougainville), New Guinea, and western Europe (Supplementary Fig. S6). The GLOBETROTTER (GT) analysis38 produced strong statistical support for two separate episodes of admixture involving the ancestors of the Leeward Society Islanders (Fig. 3, Supplementary Table S8). The first represents an average contribution of ~6% western European ancestry, which is dated to 1749–1803 CE. This is consistent with documented contact during Cook’s three voyages of exploration1, which took place 1768-71, 1772-75 and 1776-80. The second episode is estimated to have occurred in an interval from ~1,200 to 1,700 BP (229–725 CE), and is composed of a minor component (~17%), comprising mainly northern Melanesian and New Guinea sources, and a major one (~83%), in which the largest contributions are attributable to the ‘Philippines 1’, Sulawesi, and Malaysian clusters. The chronology indicates that this episode occurred prior to the earliest widely accepted radiometric dates for the permanent settlement of eastern Polynesia, which centre on ~950 BP and come from archaeological sites on Rai’atea in the Leeward Society Isles26,27. In addition, the presence of northern Melanesian ancestry in the minor component of the second (older) episode of admixture (~8% of the genome) reflects some genetic contact with this region for the ancestors of the Leeward Society Islanders prior to 1,200–1,700 BP.