|
|
Post by Admin on Jun 8, 2020 7:27:56 GMT
Genetic Distance The results of the AMOVA and MDS tests (Table 1) confirmed that the Jats had genetic affinities with several foreign populations and provided some insights into their genetic makeup. The genetic difference between the Jats and the tested populations ranged from a small distance of 0.0257 in Afghanistan to a larger distance of 0.2128 in FYR Macedonia. TABLE 1 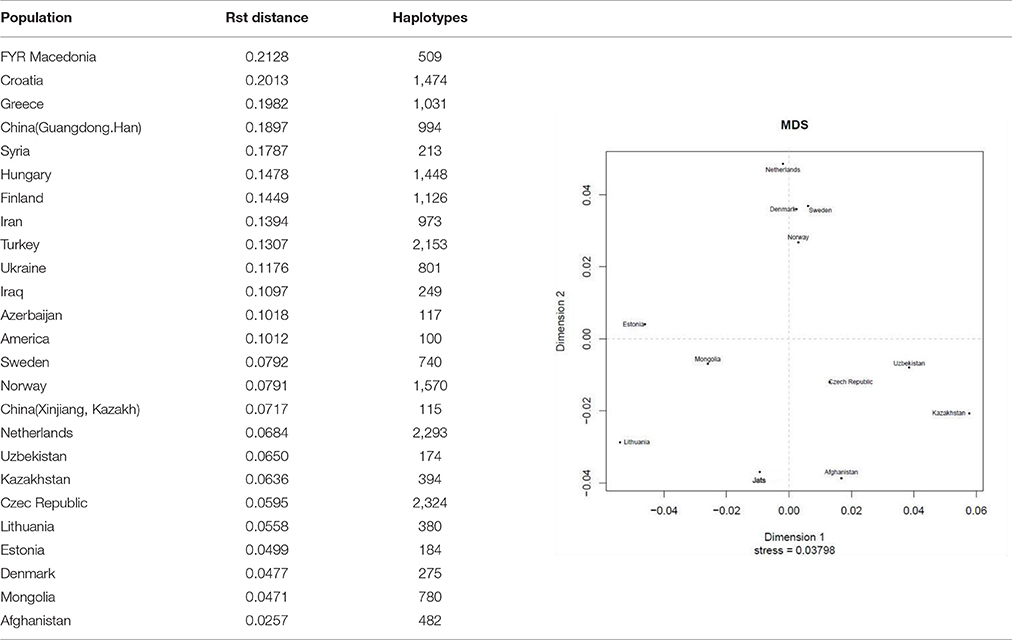 Table 1. Analysis of molecular variance (AMOVA): Pairwise Rst genetic distance between Jats and 25 selected population and a multidimensional scaling (MDS) plot of Jats and 11 closely related populations. The close genetic affinity between the Jats and the Afghani population was evident because most migrations and invasions into north India have passed through this territory. In the past, several Jat tribes and clans have inhabited parts of Afghanistan (Bellew, 1891). Haplogroups and Geographic Origins The results of haplogroup analyses revealed that MRCAs of 302 Jats in our dataset belonged to nine different haplogroups—E, G, H, I, J, L, Q, R, and T—with nine different geographic origins. The same nine haplogroups were used to compare the Jats with other ethnic groups of the Indian subcontinent. The haplogroups of 302 Jats and 1,855 other men in 38 ethnic groups of Bangladesh, India, and Pakistan are displayed in Table 2. TABLE 2 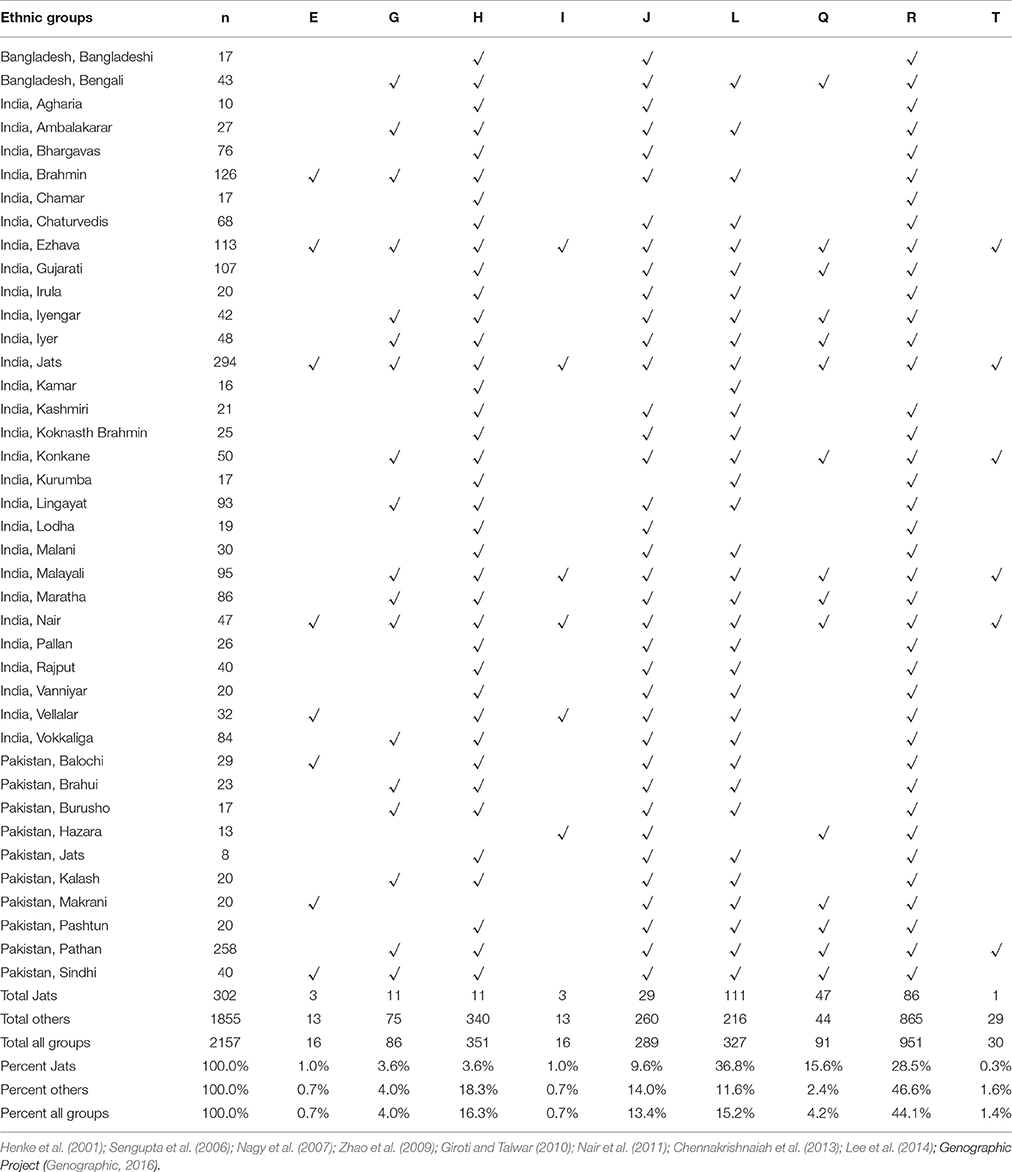 Table 2. Representation in nine Y-Chromosome haplogroups: Jats and thirty-eight ethnic groups of Indian subcontinent. The results signified that the Jats shared an underlying genetic unity with several other ethnic communities in the Indian subcontinent with the same MRCAs and geographic origins. About 90% of the Jats and about 75% of the other 38 groups in the study belonged to the same four haplogroups J, L, Q, and R. The geographic origins of the Jats in our study are summarized in Table 3. TABLE 3 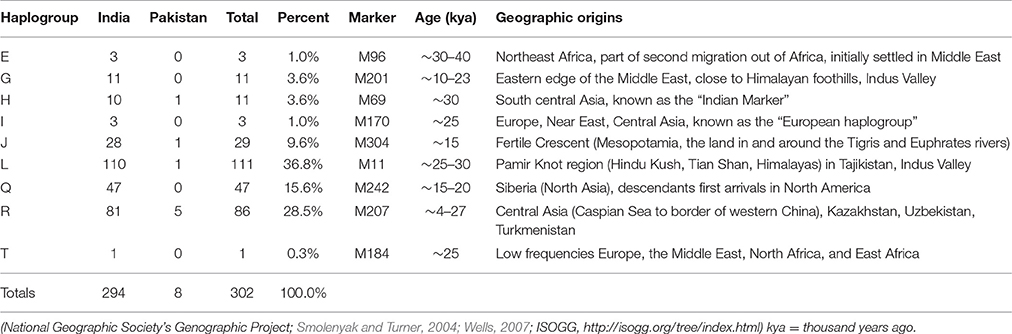 Table 3. Ancestral geographic origins of 9 Y-chromosome haplogroups of the Jats. A short phylogenetic tree of nine haplogroups of the Jats in this study—with their key top-level markers starting from Y-Adam—appears in Figure 1. FIGURE 1 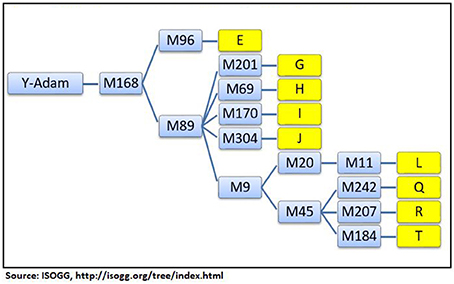 Figure 1. Phylogenetic tree of 9 Y-chromosome haplogroups of the Jats. Haplogroup L (36.8%) This is the largest haplogroup in the Jat sample population. It is present in the Indian population at an overall frequency of about 7–15% (Basu et al., 2003; Cordaux et al., 2004). Genetic studies suggest that this may be one of the original haplogroups of the creators of Indus Valley Civilization (McElreavey and Quintana-Murci, 2005; Sengupta et al., 2006). It has a frequency of about 28% in western Pakistan and Baluchistan, from where the agricultural creators of this civilization emerged (Qamar et al., 2002). The origins of this haplogroup can be traced to the rugged and mountainous Pamir Knot region in Tajikistan (Wells, 2007). Haplogroup R (28.5%) This haplogroup originated in north Asia about 27,000 years ago (ISOGG, 2017). It is one of the most common haplogroups in Europe, with its branches reaching 80% of the population in some regions. One branch is believed to have originated in the Kurgan culture, known to be the first speakers of the Indo-European languages and responsible for the domestication of the horse (Smolenyak and Turner, 2004). From somewhere in central Asia, some descendants of the man carrying the M207 mutation on the Y chromosome headed south to arrive in India about 10,000 years ago (Wells, 2007). This is one of the largest haplogroups in India and Pakistan. Of its key subclades, R2 is observed especially in India and central Asia. Haplogroup Q (15.6%) With its origins in central Asia, descendants of this group are linked to the Huns, Mongols, and Turkic people. In Europe it is found in southern Sweden, among Ashkenazi Jews, and in central and Eastern Europe such as, the Rhône-Alpes region of France, southern Sicily, southern Croatia, northern Serbia, parts of Poland and Ukraine. A subclade of this haplogroup is associated with Native American populations, and the mutation occurred 8 to 12 thousand years ago during the migration to the Americas through the Bering Strait (Smolenyak and Turner, 2004). It is estimated that as few as twenty people may have founded the initial native population of the Americas (Liu, 2016). Haplogroup J (9.6%) The ancestor of this haplogroup was born in the Middle East area known as the Fertile Crescent, comprising Israel, the West Bank, Jordon, Lebanon, Syria, and Iraq. Middle Eastern traders brought this genetic marker to the Indian subcontinent (Kerchner, 2013). Haplogroups E, G, H, I, T (9.5%) The ancestors of the remaining five haplogroups E, G, H, I, and T can be traced to different parts of Africa, Middle East, South Central Asia, and Europe (ISOGG, 2016). |
|
|
|
Post by Admin on Jun 8, 2020 20:35:26 GMT
Discussion
Sample Size
In statistical analyses, as the population increases in size, the sample size increases at a diminishing rate, and remains relatively constant when it reaches a size of 380 or more. At about 384, the sample is generally representative for a population of one million, or more (Krejcie and Morgan, 1970). Ideally, the sample size should be 380, and preferably larger.
The dataset of 302 Jats used in our research represents a margin of error of 5.7% at a confidence level of 95%. In other words, if a survey is conducted one hundred times among a similar group of people (i.e., 302 × 100; 32,000 people in total), the distribution in haplogroups is expected to be about the same as in this study, with a margin of error of plus or minus 5.7%.
Although the sample of 302 records used in this research revealed key haplogroups for the Jats, the results are not representative of this entire ethnic group of an estimated 123 million people. It is already noted that the Muslim Jats of Pakistan were underrepresented in this study. A larger sample of Muslim Jats is likely to reveal a few additional haplogroups and provide a more complete picture. Therefore, to ascertain a representative distribution of haplogroups for the entire ethnic group of the Jats, the sample size should be at least 380, with a proportional representation of Hindus, Sikhs, and Muslims.
Potential Errors in Haplogroup Prediction
Because of the need for precision in matters relating to criminal and civil laws, the forensic genetics community is generally not in favor of determining haplogroups with STR profiles. It is held that STR haplotypes are not always identical by descent, but also identical by state, and can be rooted in different haplogroups.
A study that used STR profiles of 119 males in Argentina to determine haplogroups with two software programs—Whit Athey's Haplogroup Predictor (used in this study), and a Haplogroup Classifier developed at the University of Arizona—showed that the results were not totally accurate (Muzzio et al., 2011). Another study of 165 males in Nicaragua showed that Athey's Haplogroup Predictor produced accurate results for 95.2% of the sample, but 4.8% of the results were inaccurate (Nunez et al., 2012). For greater reliability in identifying Y chromosomal haplogroups, the forensic community's preferred method is to analyze (SNPs) on the Y chromosome in the lab with actual DNA samples.
Athey has explained that the main drawback of the haplogroup prediction method in his software is the size of the database of some Y-STR haplotypes from which the allele frequencies are calculated. For most haplogroups there is sufficient Y-STR haplotype data. However, for some haplogroups, such as, C, H, L, N, and Q, the database of Y-STR haplotypes is smaller, and the results may be prone to error (Athey, 2006).
Of the 302 records used in this study, 258 were processed through Whit Athey's software. Of these, 169 haplotypes belonged to the potentially error-prone haplogroups H, L, and Q, identified by Athey. Assuming an error rate of 5% for this software, as reported in the Nicaraguan study (Nunez et al., 2012), only 13 haplotypes (5% of 258) may have identified incorrect haplogroups, representing a potential error rate of about 4.3% (13/302) in the total sample used in this study. This suggests an accuracy of about 96% in the haplogroups and geographic origins identified in this study.
Population Mixture Leading to Endogamy
Studies have shown that most ethnic groups of the Indian subcontinent descended from a mixture of two divergent populations. These were the Ancestral North Indians (ANI) who were related to Central Asians, Middle Easterners, Caucasians, and Europeans, and the Ancestral South Indians (ASI) who were not closely related to any groups outside the subcontinent (Reich et al., 2009). These findings explain that admixture was widespread at one time (Moorjani et al., 2013).
The results of the AMOVA and MDS tests in our study confirmed that the Jats had genetic contributions from several populations in the Middle East, Central Asia, and Europe. After the arrival of people called Indo-Aryans—also known as Indo-European speakers—in north India about 2000 BCE, the caste system was introduced, and a stratified social hierarchy evolved. The upper-caste populations started practicing and encouraging endogamy about 70 generations (more than 2,000 years) ago (Basu et al., 2016). Another study suggested that endogamy started much later, about the time of foreign invasions in north India (Vadivelu, 2016).
Consanguinity is another form of endogamy. The word consanguinity comes from the Latin con, meaning shared, and sanguis, meaning blood. Marriage between people who have at least one recent common ancestor is known as consanguineous, and the children are considered inbred. Couples related as second cousins or closer account for an estimated 10.4% of the global population, with the highest rates in West, Central, and South Asia (Bittles and Black, 2010). According to the International Institute for Population Sciences in Mumbai, about 16% of marriages in India are consanguineous (Kuntla et al., 2013). In Pakistan, where first cousin marriages have occurred for generations, the rate is 67% (Yaqoob et al., 1993).
The motivation behind consanguinity is usually to keep bonds, wealth, and property within a family. For this reason, there is a long list of cousin marriages among famous people (e.g., Albert Einstein, Charles Darwin, and others), and in royal families all over the world. Although endogamy has become the general norm in India, and consanguinity is practiced in some parts of the subcontinent, most ethnic groups—including the Jats—carry a blend of genetic components from different populations in the past.
Languages and Genetic Diversity
There are several thousand ethnic and tribal groups in the Indian subcontinent (Papiha, 1996; Xing et al., 2010). Members of these communities share common self-identities that are based on languages, customs, cuisines, and at least six major religions. There are 22 official languages and many dialects in the country (Annamalai, 2006). At least eight different languages—Balochi, Haryanvi, Hindi, Punjabi, Rajasthani, Saraiki, Sindhi, and Urdu—are spoken in the Jat communities, which demonstrates their genetic diversity.
The Aryan-Scythian Conundrum
The estimated population of the Indian subcontinent in 10,000 BCE was about 100,000 people, and stayed at this level until about 5,000 BCE, by when agriculture had spread in the Indus Valley (McEvedy and Jones, 1978). Since then the population has grown exponentially, with about 1.7 billion people in the Indian subcontinent now. Among the several thousand ethnic and tribal groups in the subcontinent, there are no existing population groups known as Indo-Aryan or Indo-Scythian. These appear to be labels that have been loosely applied to people who arrived in north India in a series of waves over a long period in the distant past.
Sir Risley's ethnographic classifications of Indian people did not provide any clues about the origins of the Indo-Aryans and the Scytho-Dravidians. But his studies showed that these two groups were physically different. According to the Imperial Gazetteer of India, the Indo-Scythians were likely pushed toward the south by the Indo-Aryans, mingled with the Dravidian population, and became the ancestors of an entirely different ethnic group known as the Marathas (Gazetteer, 1931).
The Pamir Knot region—from where the MRCA of haplogroup L emerged—is also the home of the Bactria-Margiana Archaeological Complex (BMAC), in a site called Gonur that represents a Bronze Age culture known as the Oxus civilization (Sarianidi, 2007). This BMAC site of around 4000 BCE was discovered and named by the Soviet archeologist Viktor Sarianidi. Among his findings, Sarianidi discovered evidence of sacred alters; traces of ingredients such as, poppy seeds, cannabis, and ephedra, used for a drink called soma; horse sacrifices; four wheeled chariots; and other connections with the Aryans (Sarianidi, 2007; Wood, 2007). Some BMAC materials of this type have been found in the Indus Valley sites. Archaeologist J. P. Mallory from Queens University (Ireland), and Indologist Asko Parpola from the University of Helsinki (Finland), have suggested a connection between the Aryans and BMAC (Mallory, 1989; Parpola, 1999). Because the MRCA of haplogroup L emerged from the same geographical area as the people called the Aryans, there may be a genetic link between the two.
The haplotypes of 26 ancient human specimens from the Krasnoyarsk area in Siberia, dated from between the middle of the second millennium BCE to the fourth century CE (Scythian and Sarmatian timeframe), revealed that nearly all specimens belonged to R1a, a subclade of haplogroup R, which is thought to mark the eastward migration of early Indo-Europeans (Keyser et al., 2009). Another survey of 217 samples from Europe and Asia revealed that R1a1, another subclade of haplogroup R, was spread across Eurasia (Pamjav et al., 2012). Because the origins of haplogroup R can be traced to the same geographical area, there may be a genetic link with the ancient people called Scythians.
Studies have shown that the Hindu Kush area from where these groups migrated to the Indian subcontinent served as a confluence of gene flows from adjoining areas rather than a source of distinctly autochthonous populations (Cristofaro et al., 2013). These people also arrived in north India at different times. As noted earlier, members of haplogroup R arrived about 10,000 years ago, the Indo-Aryan migrations started about 2000 BCE, and the Indo-Scythians arrived much later, around 200 BCE. Because of their physical differences and the large gaps between their arrival times, it can be inferred that these groups were genetically different and not the same people.
This study has shown that the genetic origins of the Jats can be traced to at least nine and possibly more MRCA's, with nine different geographical origins that are spread thousands of miles apart (e.g., from the Fertile Crescent to Serbia). These nine MRCAs were genetically different. Therefore, any assertion that Jats are descendants of a single ancient population such as, the Indo-Aryans or Indo-Scythians cannot be supported. However, certain members of the Jat ethnic group who belong to haplogroups L and R—along with members of several other ethnic groups in the Indian subcontinent who belong to the same two haplogroups—are the most probable candidates to be linked to these ancient populations.
Conclusion
The human Y-chromosome provides a powerful molecular tool for analyzing Y-STR haplotypes and determining their haplogroups which lead to the ancient geographic origins of individuals. For this study, the Jats and 38 other ethnic groups in the Indian subcontinent were analyzed, and their haplogroups were compared. Using genetic markers and available descriptions of haplogroups from the Y-DNA phylogenetic tree, the geographic origins and migratory paths of their ancestors were traced.
The study demonstrated that based on their genetic makeup, the Jats belonged to at least nine specific haplogroups, with nine different lines of ancestry and geographic origins. About 90% of the Jats in our sample belonged to only four different lines of ancestry and geographic origins. Therefore, attributing the origins of this entire ethnic group to loosely defined ancient populations such as, Indo-Aryans or Indo-Scythians represents very broad generalities and cannot be supported. The study also revealed that even with their different languages, religions, nationalities, customs, cuisines, and physical differences, the Jats shared their haplogroups with several other ethnic groups of the Indian subcontinent, and had the same common ancestors and geographic origins in the distant past. Based on recent developments in DNA science, this study provided new insights into the ancient geographic origins of this major ethnic group in the Indian subcontinent. A larger dataset, particularly with more representation of Muslim Jats, is likely to reveal some additional haplogroups and geographical origins for this ethnic group.
|
|
|
|
Post by Admin on Mar 28, 2021 19:27:34 GMT
#thinkschoolenglish #imtiazshams #থিংকইংলিশ How Ancient DNA is Rewriting India’s History | Think English How Ancient DNA is Rewriting India’s History - New DNA research on the Indus Valley civilisation is changing what we know about the subcontinent’s early people. That affects modern people, too. --------- We're a charity that makes videos on science, history and culture in many languages. Support our work by becoming a patron on Patreon: www.patreon.com/thinkcharity |
|
|
|
Post by Admin on Dec 27, 2021 20:42:04 GMT
The thorniest, most fought-over question in Indian history is slowly but surely getting answered: did Indo-European language speakers, who called themselves Aryans, stream into India sometime around 2,000 BC – 1,500 BC when the Indus Valley civilisation came to an end, bringing with them Sanskrit and a distinctive set of cultural practices? Genetic research based on an avalanche of new DNA evidence is making scientists around the world converge on an unambiguous answer: yes, they did. This may come as a surprise to many — and a shock to some — because the dominant narrative in recent years has been that genetics research had thoroughly disproved the Aryan migration theory. This interpretation was always a bit of a stretch as anyone who read the nuanced scientific papers in the original knew. But now it has broken apart altogether under a flood of new data on Y-chromosomes (or chromosomes that are transmitted through the male parental line, from father to son). Lines of descent Until recently, only data on mtDNA (or matrilineal DNA, transmitted only from mother to daughter) were available and that seemed to suggest there was little external infusion into the Indian gene pool over the last 12,500 years or so. New Y-DNA data has turned that conclusion upside down, with strong evidence of external infusion of genes into the Indian male lineage during the period in question. The reason for the difference in mtDNA and Y-DNA data is obvious in hindsight: there was strong sex bias in Bronze Age migrations. In other words, those who migrated were predominantly male and, therefore, those gene flows do not really show up in the mtDNA data. On the other hand, they do show up in the Y-DNA data: specifically, about 17.5% of Indian male lineage has been found to belong to haplogroup R1a (haplogroups identify a single line of descent), which is today spread across Central Asia, Europe and South Asia. Pontic-Caspian Steppe is seen as the region from where R1a spread both west and east, splitting into different sub-branches along the way. The paper that put all of the recent discoveries together into a tight and coherent history of migrations into India was published just three months ago in a peer-reviewed journal called ‘BMC Evolutionary Biology’. In that paper, titled “A Genetic Chronology for the Indian Subcontinent Points to Heavily Sex-biased Dispersals”, 16 scientists led by Prof. Martin P. Richards of the University of Huddersfield, U.K., concluded: “Genetic influx from Central Asia in the Bronze Age was strongly male-driven, consistent with the patriarchal, patrilocal and patrilineal social structure attributed to the inferred pastoralist early Indo-European society. This was part of a much wider process of Indo-European expansion, with an ultimate source in the Pontic-Caspian region, which carried closely related Y-chromosome lineages… across a vast swathe of Eurasia between 5,000 and 3,500 years ago”. In an email exchange, Prof. Richards said the prevalence of R1a in India was “very powerful evidence for a substantial Bronze Age migration from central Asia that most likely brought Indo-European speakers to India.” The robust conclusions of Professor Richards and his team rest on their own substantive research as well as a vast trove of new data and findings that have become available in recent years, through the work of genetic scientists around the world. What’s happened very rapidly, dramatically, and powerfully in the last few years has been the explosion of genome-wide studies of human history based on modern and ancient DNA, and that’s been enabled by the technology of genomics and the technology of ancient DNA....” David Reich, Geneticist and professor, Harvard Medical School. 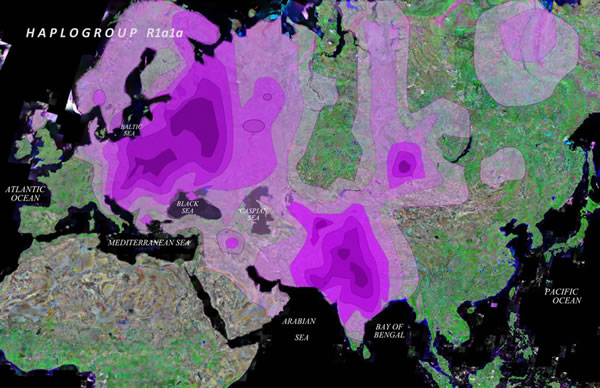 Peter Underhill, scientist at the Department of Genetics at the Stanford University School of Medicine, is one of those at the centre of the action. Three years ago, a team of 32 scientists he led published a massive study mapping the distribution and linkages of R1a. It used a panel of 16,244 male subjects from 126 populations across Eurasia. Dr. Underhill’s research found that R1a had two sub-haplogroups, one found primarily in Europe and the other confined to Central and South Asia. Ninety-six per cent of the R1a samples in Europe belonged to sub-haplogroup Z282, while 98.4% of the Central and South Asian R1a lineages belonged to sub-haplogroup Z93. The two groups diverged from each other only about 5,800 years ago. Dr. Underhill’s research showed that within the Z93 that is predominant in India, there is a further splintering into multiple branches. The paper found this “star-like branching” indicative of rapid growth and dispersal. So if you want to know the approximate period when Indo-European language speakers came and rapidly spread across India, you need to discover the date when Z93 splintered into its own various subgroups or lineages. We will come back to this later. So in a nutshell: R1a is distributed all over Europe, Central Asia and South Asia; its sub-group Z282 is distributed only in Europe while another subgroup Z93 is distributed only in parts of Central Asia and South Asia; and three major subgroups of Z93 are distributed only in India, Pakistan, Afghanistan and the Himalayas. This clear picture of the distribution of R1a has finally put paid to an earlier hypothesis that this haplogroup perhaps originated in India and then spread outwards. This hypothesis was based on the erroneous assumption that R1a lineages in India had huge diversity compared to other regions, which could be indicative of its origin here. As Prof. Richards puts it, “the idea that R1a is very diverse in India, which was largely based on fuzzy microsatellite data, has been laid to rest” thanks to the arrival of large numbers of genomic Y-chromosome data. |
|
|
|
Post by Admin on Dec 27, 2021 21:10:09 GMT
Gene-dating the migration
Now that we know that there WAS indeed a significant inflow of genes from Central Asia into India in the Bronze Age, can we get a better fix on the timing, especially the splintering of Z93 into its own sub-lineages? Yes, we can; the research paper that answers this question was published just last year, in April 2016, titled: “Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences.” This paper, which looked at major expansions of Y-DNA haplogroups within five continental populations, was lead-authored by David Poznik of the Stanford University, with Dr. Underhill as one of the 42 co-authors. The study found “the most striking expansions within Z93 occurring approximately 4,000 to 4,500 years ago”. This is remarkable, because roughly 4,000 years ago is when the Indus Valley civilization began falling apart. (There is no evidence so far, archaeologically or otherwise, to suggest that one caused the other; it is quite possible that the two events happened to coincide.)
The avalanche of new data has been so overwhelming that many scientists who were either sceptical or neutral about significant Bronze Age migrations into India have changed their opinions. Dr. Underhill himself is one of them. In a 2010 paper, for example, he had written that there was evidence “against substantial patrilineal gene flow from East Europe to Asia, including to India” in the last five or six millennia. Today, Dr. Underhill says there is no comparison between the kind of data available in 2010 and now. “Then, it was like looking into a darkened room from the outside through a keyhole with a little torch in hand; you could see some corners but not all, and not the whole picture. With whole genome sequencing, we can now see nearly the entire room, in clearer light.”
Dr. Underhill is not the only one whose older work has been used to argue against Bronze Age migrations by Indo-European language speakers into India. David Reich, geneticist and professor in the Department of Genetics at the Harvard Medical School, is another one, even though he was very cautious in his older papers. The best example is a study lead-authored by Reich in 2009, titled “Reconstructing Indian Population History” and published in Nature. This study used the theoretical construct of “Ancestral North Indians” (ANI) and “Ancestral South Indians” (ASI) to discover the genetic substructure of the Indian population. The study proved that ANI are “genetically close to Middle Easterners, Central Asians, and Europeans”, while the ASI were unique to India. The study also proved that most groups in India today can be approximated as a mixture of these two populations, with the ANI ancestry higher in traditionally upper caste and Indo-European speakers. By itself, the study didn’t disprove the arrival of Indo-European language speakers; if anything, it suggested the opposite, by pointing to the genetic linkage of ANI to Central Asians.
However, this theoretical structure was stretched beyond reason and was used to argue that these two groups came to India tens of thousands of years ago, long before the migration of Indo-European language speakers that is supposed to have happened only about 4,000 to 3,500 years ago. In fact, the study had included a strong caveat that suggested the opposite: “We caution that ‘models’ in population genetics should be treated with caution. While they provide an important framework for testing historical hypothesis, they are oversimplifications. For example, the true ancestral populations were probably not homogenous as we assume in our model but instead were likely to have been formed by clusters of related groups that mixed at different times.” In other words, ANI is likely to have resulted from multiple migrations, possibly including the migration of Indo-European language speakers.
The spin and the facts
But how was this research covered in the media? “Aryan-Dravidian divide a myth: Study,” screamed a newspaper headline on September 25, 2009. The article quoted Lalji Singh, a co-author of the study and a former director of the Centre for Cellular and Molecular Biology (CCMB), Hyderabad, as saying: “This paper rewrites history… there is no north-south divide”. The report also carried statements such as: “The initial settlement took place 65,000 years ago in the Andamans and in ancient south India around the same time, which led to population growth in this part. At a later stage, 40,000 years ago, the ancient north Indians emerged which in turn led to rise in numbers there. But at some point in time, the ancient north and the ancient south mixed, giving birth to a different set of population. And that is the population which exists now and there is a genetic relationship between the population within India.” The study, however, makes no such statements whatsoever — in fact, even the figures 65,000 and 40,000 do not figure it in it!
This stark contrast between what the study says and what the media reports said did not go unnoticed. In his column for Discover magazine, geneticist Razib Khan said this about the media coverage of the study: “But in the quotes in the media the other authors (other than Reich that is - ed) seem to be leading you to totally different conclusions from this. Instead of leaning toward ANI being proto-Indo-European, they deny that it is.”
Let’s leave that there, and ask what Reich says now, when so much new data have become available? In an interview with Edge in February last year, while talking about the thesis that Indo-European languages originated in the Steppes and then spread to both Europe and South Asia, he said: “The genetics is tending to support the Steppe hypothesis because in the last year, we have identified a very strong pattern that this ancient North Eurasian ancestry that you see in Europe today, we now know when it arrived in Europe. It arrived 4500 years ago from the East from the Steppe...” About India, he said: “In India, you can see, for example, that there is this profound population mixture event that happens between 2000 to 4000 years ago. It corresponds to the time of the composition of the Rigveda, the oldest Hindu religious text, one of the oldest pieces of literature in the world, which describes a mixed society...” In essence according to Reich, in broadly the same time frame, we see Indo-European language speakers spreading out both to Europe and to South Asia, causing major population upheavals.
The dating of the “profound population mixture event” that Reich refers to was arrived at in a paper that was published in the American Journal of Human Genetics in 2013, and was lead authored by Priya Moorjani of the Harvard Medical School, and co-authored, among others, by Reich and Lalji Singh. This paper too has been pushed into serving the case against migrations of Indo-European language speakers into India, but the paper itself says no such thing, once again!
Here’s what it says in one place: “The dates we report have significant implications for Indian history in the sense that they document a period of demographic and cultural change in which mixture between highly differentiated populations became pervasive before it eventually became uncommon. The period of around 1,900–4,200 years before present was a time of profound change in India, characterized by the de-urbanization of the Indus civilization, increasing population density in the central and downstream portions of the Gangetic system, shifts in burial practices, and the likely first appearance of Indo-European languages and Vedic religion in the subcontinent.”
The study didn’t “prove” the migration of Indo-European language speakers since its focus was different: finding the dates for the population mixture. But it is clear that the authors think its findings fit in well with the traditional reading of the dates for this migration. In fact, the paper goes on to correlate the ending of population mixing with the shifting attitudes towards mixing of the races in ancient texts. It says: “The shift from widespread mixture to strict endogamy that we document is mirrored in ancient Indian texts.”
So irrespective of the use to which Priya Moorjani et al’s 2013 study is put, what is clear is that the authors themselves admit their study is fully compatible with, and perhaps even strongly suggests, Bronze Age migration of Indo-European language speakers. In an email to this writer, Moorjani said as much. In answer to a question about the conclusions of the recent paper of Prof. Richards et al that there were strong, male-driven genetic inflows from Central Asia about 4,000 years ago, she said she found their results “to be broadly consistent with our model”. She also said the authors of the new study had access to ancient West Eurasian samples “that were not available when we published in 2013”, and that these samples had provided them additional information about the sources of ANI ancestry in South Asia.
One by one, therefore, every single one of the genetic arguments that were earlier put forward to make the case against Bronze Age migrations of Indo-European language speakers have been disproved. To recap:
1. The first argument was that there were no major gene flows from outside to India in the last 12,500 years or so because mtDNA data showed no signs of it. This argument was found faulty when it was shown that Y-DNA did indeed show major gene flows from outside into India within the last 4000 to 4,500 years or so, especially R1a which now forms 17.5% of the Indian male lineage. The reason why mtDNA data behaved differently was that Bronze Age migrations were severely sex-biased.
2. The second argument put forward was that R1a lineages exhibited much greater diversity in India than elsewhere and, therefore, it must have originated in India and spread outward. This has been proved false because a mammoth, global study of R1a haplogroup published last year showed that R1a lineages in India mostly belong to just three subclades of the R1a-Z93 and they are only about 4,000 to 4,500 years old.
3. The third argument was that there were two ancient groups in India, ANI and ASI, both of which settled here tens of thousands of years earlier, much before the supposed migration of Indo-European languages speakers to India. This argument was false to begin with because ANI — as the original paper that put forward this theoretical construct itself had warned — is a mixture of multiple migrations, including probably the migration of Indo-European language speakers.
Connecting the dots
Two additional things should be kept in mind while looking at all this evidence. The first is how multiple studies in different disciplines have arrived at one specific period as an important marker in the history of India: around 2000 B.C. According to the Priya Moorjani et al study, this is when population mixing began on a large scale, leaving few population groups anywhere in the subcontinent untouched. The Onge in the Andaman and Nicobar Islands are the only ones we know to have been completely unaffected by what must have been a tumultuous period. And according to the David Poznik et al study of 2016 on the Y-chromosome, 2000 B.C. is around the time when the dominant R1a subclade in India, Z93, began splintering in a “most striking” manner, suggesting “rapid growth and expansion”. Lastly, from long-established archaeological studies, we also know that 2000 BC was around the time when the Indus Valley civilization began to decline. For anyone looking at all of these data objectively, it is difficult to avoid the feeling that the missing pieces of India’s historical puzzle are finally falling into place.
The second is that many studies mentioned in this piece are global in scale, both in terms of the questions they address and in terms of the sampling and research methodology. For example, the Poznik study that arrived at 4,000-4,500 years ago as the dating for the splintering of the R1a Z93 lineage, looked at major Y-DNA expansions not just in India, but in four other continental populations. In the Americas, the study proved the expansion of haplogrop Q1a-M3 around 15,000 years ago, which fits in with the generally accepted time for the initial colonisation of the continent. So the pieces that are falling in place are not merely in India, but all across the globe. The more the global migration picture gets filled in, the more difficult it will be to overturn the consensus that is forming on how the world got populated.
Nobody explains what is happening now better than Reich: “What’s happened very rapidly, dramatically, and powerfully in the last few years has been the explosion of genome-wide studies of human history based on modern and ancient DNA, and that’s been enabled by the technology of genomics and the technology of ancient DNA. Basically, it’s a gold rush right now; it’s a new technology and that technology is being applied to everything we can apply it to, and there are many low-hanging fruits, many gold nuggets strewn on the ground that are being picked up very rapidly.”
So far, we have only looked at the migrations of Indo-European language speakers because that has been the most debated and argued about historical event. But one must not lose the bigger picture: R1a lineages form only about 17.5 % of Indian male lineage, and an even smaller percentage of the female lineage. The vast majority of Indians owe their ancestry mostly to people from other migrations, starting with the original Out of Africa migrations of around 55,000 to 65,000 years ago, or the farming-related migrations from West Asia that probably occurred in multiple waves after 10,000 B.C., or the migrations of Austro-Asiatic speakers such as the Munda from East Asia the dating of which is yet to determined, and the migrations of Tibeto-Burman speakers such as the Garo again from east Asia, the dating of which is also yet to be determined.
What is abundantly clear is that we are a multi-source civilization, not a single-source one, drawing its cultural impulses, its tradition and practices from a variety of lineages and migration histories. The Out of Africa immigrants, the pioneering, fearless explorers who discovered this land originally and settled in it and whose lineages still form the bedrock of our population; those who arrived later with a package of farming techniques and built the Indus Valley civilization whose cultural ideas and practices perhaps enrich much of our traditions today; those who arrived from East Asia, probably bringing with them the practice of rice cultivation and all that goes with it; those who came later with a language called Sanskrit and its associated beliefs and practices and reshaped our society in fundamental ways; and those who came even later for trade or for conquest and chose to stay, all have mingled and contributed to this civilization we call Indian. We are all migrants.
|
|









