|
|
Post by Admin on Jan 26, 2016 6:35:27 GMT
 The Brahmin is a member of the highest of the four Indian castes, who teaches Vedic literature (Brahmin, the priests; Kshatriya, warriors and nobility; Vaisya, farmers, traders and artisans; and Shudra, tenant farmers and servants.) The Indo-Aryan people migrated to northwestern parts of the Indian subcontinent and spread to the Ganges Plain around 1200 BCE, which coincided with the spread of the Vedic culture in India. Brahmin people are genetically distinct from other Indians outside the Brahmin caste. There is a high percentage of blue eyes among the Brahmin caste and the highest levels of Haplogroup R1a are found among West Bengal Brahmins (72%) and Uttar Pradesh Brahmins (67%). Moreover, a previous genetic study concluded that the Indian upper castes are significantly more similar to Europeans than the lower castes. The degree of West Eurasian admixture was proportional to caste rank as more West Eurasians became members of the upper castes at the inception of the caste system in India (Bamshad et al. 2001).  The origins and affinities of the ∼1 billion people living on the subcontinent of India have long been contested. This is owing, in part, to the many different waves of immigrants that have influenced the genetic structure of India. In the most recent of these waves, Indo-European-speaking people from West Eurasia entered India from the Northwest and diffused throughout the subcontinent. They purportedly admixed with or displaced indigenous Dravidic-speaking populations. Subsequently they may have established the Hindu caste system and placed themselves primarily in castes of higher rank. To explore the impact of West Eurasians on contemporary Indian caste populations, we compared mtDNA (400 bp of hypervariable region 1 and 14 restriction site polymorphisms) and Y-chromosome (20 biallelic polymorphisms and 5 short tandem repeats) variation in ∼265 males from eight castes of different rank to ∼750 Africans, Asians, Europeans, and other Indians. For maternally inherited mtDNA, each caste is most similar to Asians. However, 20%–30% of Indian mtDNA haplotypes belong to West Eurasian haplogroups, and the frequency of these haplotypes is proportional to caste rank, the highest frequency of West Eurasian haplotypes being found in the upper castes. In contrast, for paternally inherited Y-chromosome variation each caste is more similar to Europeans than to Asians. 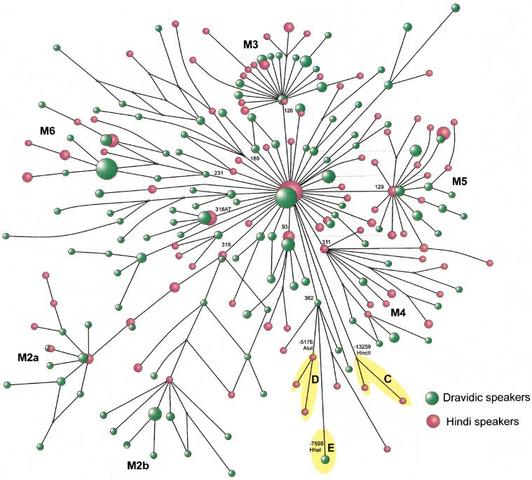 Phylogeny of haplogroup M in India. Moreover, the affinity to Europeans is proportionate to caste rank, the upper castes being most similar to Europeans, particularly East Europeans. These findings are consistent with greater West Eurasian male admixture with castes of higher rank. Nevertheless, the mitochondrial genome and the Y chromosome each represents only a single haploid locus and is more susceptible to large stochastic variation, bottlenecks, and selective sweeps. Thus, to increase the power of our analysis, we assayed 40 independent, biparentally inherited autosomal loci (1 LINE-1 and 39 Alu elements) in all of the caste and continental populations (∼600 individuals). Analysis of these data demonstrated that the upper castes have a higher affinity to Europeans than to Asians, and the upper castes are significantly more similar to Europeans than are the lower castes. Collectively, all five datasets show a trend toward upper castes being more similar to Europeans, whereas lower castes are more similar to Asians. We conclude that Indian castes are most likely to be of proto-Asian origin with West Eurasian admixture resulting in rank-related and sex-specific differences in the genetic affinities of castes to Asians and Europeans.  Affinities to Europeans and Asians Stratified by Caste Rank Genetic distances estimated from autosomal Alu elements correspond to caste rank, the genetic distance between the upper and lower castes being more than 2.5 times larger than the distance between upper and middle or middle and lower castes (upper to middle, 0.0069; upper to lower, 0.018; middle to lower, 0.0071). These trends are the same whether the Kshatriya and Vysya are included in the upper castes, the middle castes, or excluded from the analysis (data not shown). Furthermore, a neighbor-joining network of genetic distances between separate castes (Fig. (Fig.3)3) clearly differentiates castes of different rank into separate clusters. This is similar to the relationship between genetic distances and caste rank estimated from mtDNA (Bamshad et al. 1998). It is important to note, however, that the autosomal genetic distances are estimated from 40 independent loci. This afforded us the opportunity to test the statistical significance of the correspondence between genetic distance and caste status. The Mantel correlation between interindividual genetic distances and distances based on social rank was low but highly significant for individuals ranked into upper, middle, and lower groups (r = 0.08; p < 0.001) and into eight separate castes (r = 0.07; p < 0.001). Given the resolving power of this autosomal dataset, we next tested whether we could reconcile the results of the analysis of mtDNA and Y-chromosome markers in castes and continental populations. 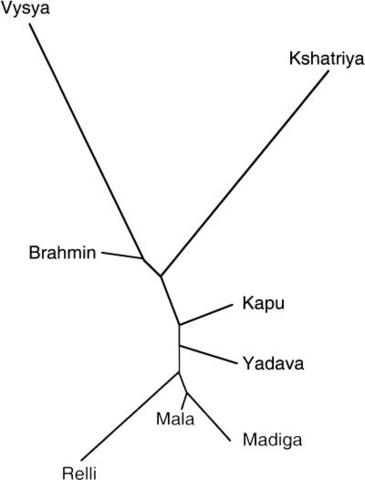 Neighbor-joining network of genetic distances among caste communities estimated from 40 Alu polymorphisms. Distances between upper castes (U; Brahmin, Vysya, Kshatriya), middle castes (M; Yadava, Kapu), and lower castes (L; Mala, Madiga, Relli) are significantly correlated with social rank. Genotypic differentiation was significantly different from zero (p < 0.0001) between each pair of caste populations and between each caste and continental population. Similar to the results of both the mtDNA and Y-chromosome analyses, the distance between upper castes and European populations is smaller than the distance between lower castes and Europeans (Table (Table5).5). However, in contrast to the mtDNA results but similar to the Y-chromosome results, the affinity between upper castes and Europeans is higher than that of upper castes and Asians (Table (Table5).5). If the Kshatriya and Vysya are excluded from the analysis or included in the middle castes, the genetic distance between the upper caste (Brahmins) and Europeans remains smaller than the distance between the lower castes and Europeans and the distance between upper castes and Asians (Table (Table5).5). Analysis of each caste separately reveals that the genetic distance between the Brahmins and Europeans (0.013) is less than the distance between Europeans and Kshatryia (0.030) or Vysya (0.020). Nevertheless, each separate upper caste is more similar to Europeans than to Asians. Because historical evidence suggests greater affinity between upper castes and Europeans than between lower castes and Europeans (Balakrishnan 1978, 1982; Cavalli-Sforza et al. 1994), it is appropriate to use a one-tailed test of the difference between the corresponding genetic distances. The 90% confidence limits of Nei's standard distances estimated between upper castes and Europeans (0.006–0.016) versus lower castes and Europeans (0.017–0.037) do not overlap, indicating statistical significance at the 0.05 level. Significance at 0.05 is not achieved if the Kshatriya and Vysya are excluded. These results offer statistical support for differences in the genetic affinity of Europeans to caste populations of differing rank, with greater European affinity to upper castes than to lower castes. Genome Res. 2001 Jun; 11(6): 994–1004. |
|
|
|
Post by Admin on May 1, 2016 22:51:31 GMT
 Tamil Bride in Antique Kundan Sets The proto-Dravidian people may have arrived from the Fertile Crescent, which explains the presence of the bull-leaping tradition shared with ancient Mesopotamians and the Minoans, who were the first Europeans to have literate civilisation. The proto-Elamo-Dravidian language originated in the Elam province in southwestern Iran, and spread eastwards to the Indus Valley and the Indian sub-continent. The proto-Dravidian people were preceded by the Tibeto-Burman language speakers, who live in six of the seven states of Northeastern India. Tibeto-Burman languages are currently spoken in Tibet and western China and the Tibeto-Burman language speakers were most likely to be the original inhabitants of the Indian sub-continent.  Although Paleolithic and Mesolithic people left their mark in the Iranian Plateau, major human population developments with possible genetic implications occurred here during the Neolithic period and later [1], [10], [11]. The Middle Eastern region spanning from Zagros Mountains and northern Mesopotamia to Southeast Anatolia, called Fertile Crescent, is broadly accepted to be the place where agriculture first arose [1]. Important agricultural developments occurred in the eastern horn of the Fertile Crescent, notably in Elam (southwestern Iran), connecting Mesopotamia and the Iranian Plateau [12]. The highly urban Elamite civilization had close contacts with Mesopotamians but exhibited an extensive differentiation from the rest of the Fertile Crescent populations, including a language that is thought to belong to the Dravidian family [3], [13]. Another major innovation, that most likely emerged later than agriculture, was the domestication of animals, which is thought to have led to dramatic population expansions in Eurasia [1], [14], [15]. Starting about 5000 years (ky) before present, pastoral nomadism developed in the grasslands of Central Asia, as well as in southeastern Europe, opening up the possibility of rapid movements of large population groups [16]. The spread of these new technologies has been associated with the dispersal of Dravidian and Indo-European languages in southern Asia [17], [18]. It is hypothesized that the proto-Elamo-Dravidian language, most likely originated in the Elam province in southwestern Iran, spread eastwards with the movement of farmers to the Indus Valley and the Indian sub-continent [13], [19]. Between the third and second millennia BCE the Iranian Plateau became exposed to incursions of pastoral nomads from the Central Asian steppes, who brought the Indo-Iranian language of the Indo-European family, which eventually replaced Dravidian languages, perhaps by an elite-dominance model [13], [17], [20].  Figure 1. MDS plot based on Fst statistics calculated from complete mtDNA sequences for population samples from Iran, Anatolia, Caucasus, and Europe. In order to visualize the relationships between Iranians studied and other populations of the Caucasus, Anatolia and Europe based on complete mtDNA sequence data, MDS plot was constructed from the pairwise Fst values (Table S3). The results show that Persians and Qashqais are close to each other and to Armenians, whereas Azeris from Iran are located nearby Georgians. It is worth pointing out the position of Azeris from the Caucasus region, who despite their supposed common origin with Iranian Azeris, cluster quite separately and occupy an intermediate position between the Azeris/Georgians and Turks/Iranians grouping (Figure 1). Interestingly, the results of our MDS analysis do not combine the populations studied according to their geographic and/or linguistic affinity. Therefore, Turkic-speaking Qashqais, Azeris, and Turks are located quite distantly from each other on the plot, even though association between the latter two groups has been recently revealed based on complete mtDNA sequences [40]. All populations from the Caucasus region (Armenians, Azeris, and Georgians) are scattered on the plot though their genetic proximity has been demonstrated by Schönberg et al. [40]. Similarly, Iranians from Tehran province [40] and Persians studied here are clearly separated from each other. The genetic structure of Iranians in comparison with the populations from the Caucasus, Anatolia and Europe was investigated by AMOVA (Table S4). As expected, before grouping, the majority of variability was due to within population component (98.5% for Iranian populations only and 98.12% for the complete data set). After grouping, neither geographic nor linguistic classification gave a good fit to the genetic data. A slightly higher degree of geographic rather than linguistic correlation with the genetic structuring of the examined populations emerges only when the geographically and genetically distant population of Buryats from Inner Mongolia region of China was added to the studied data set, thus underlining the importance of geographic distance. It should be noted that correlation between geographical proximity and genetic relationships of populations has been shown previously based on HVS1 variability data for Indo-European and Semitic-speaking groups of southwestern Iran [56], [57]. Moreover, a slightly better fit of geographic rather than linguistic classification of populations to the complete mtDNA sequence data has been demonstrated recently for Iranian, Anatolian and Caucasus region populations [40]. 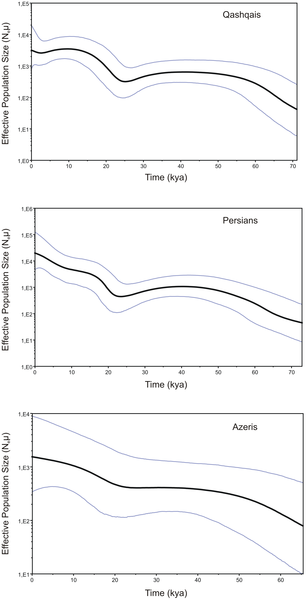 Figure 2. BSP indicating the median of the hypothetical effective population size through time based on complete mtDNA genome data for Persians, Qashqais and Azeris. The haplogroup assignment for each individual according to the nomenclature of Phylotree.org [43] (Build 15) is given in Table S4. A total of 212 different sub-haplogroups or paragroups (unclassified lineages within a clade) were identified, which fall into 75 principal haplogroups. The vast majority of the mtDNAs clustered into macrohaplogroups M, N, and R, but a limited number was found to belong to the sub-Saharan haplogroups L2a, L3d, L3e, L3f and L5c (Figure S2). Two haplogroups, H91 and HV18, are defined here for the first time, whereas others (marked red in Table S1 and Figure S2) represent newly identified sub-clades. Moreover, for some previously known haplogroups we redefined diagnostic markers that allow a better definition of the haplogroup topology within the tree. Haplogroup frequencies for Azeris, Persians, Qashqais and the entire Iranian mtDNA data set are presented in Table 2. All Iranian populations studied here are characterized by the same most prominent western Eurasian mtDNA haplogroups, H, J, T and U; however, the frequency distribution of these lineages varies between different populations. All the three populations show similar frequencies of haplogroup U (22.7%, 24.3%, and 22.3%, respectively), but the frequencies of haplogroups H and J are more pronounced in Qashqais (28.6% and 15.2%, respectively) than in Persians (16.6% and 6.1%, respectively) and Azeris (22.7% and 4.6%, respectively). In contrast, haplogroup T dominates in Azeris (18.2%) and Persians (11.6%), being found only in 4.5% in Qashqais. Another notable difference between Azeris and two other populations studied is the higher frequency of haplogroups X2 (9.1% versus 2.8% in Persians and 2.7% in Qashqais) and N1a3 (9.1% versus 0.6% in Persians), and the absence of haplogroups U and H diversification, which may be merely the result of smaller sample size. There are also distinguished differences between the Persians and Qashqais with respect to the distribution of some sub-haplogroups, including H13 (2.8% versus 6.3%), T2 (7.2% versus 1.8%), U3 (2.8% versus 8%), and U7 (7.2% versus 2.7%) (Table 2).  Table 2. Mitochondrial haplogroup frequencies (%) in Iranian populations. It is known that some of the mtDNA lineages are either autochthonous to Iran or underwent a major expansion in this region [11]. Among them is haplogroup R2, which is concentrated in southern Pakistan and in India, and is present at low frequencies in the most of adjacent regions, including the Near East, the Caucasus, the Iranian Plateau, the Arabian Peninsula, and Central Asia [11], [25], [37]. The extensive sequencing of complete mtDNAs from a large part of the Iranian Plateau led us to the identification of several highly divergent Qashqai lineages within the entire haplogroup R2 and revealed a new Persian-specific sub-clade within haplogroup R2a. The reconstructed complete mtDNA phylogeny based on fifteen published and nine new Iranian R2 sequences is shown in Figure 3 and includes age estimates obtained from complete genome and synonymous mutations. As can be seen, haplogroup R2 has a likely pre-LGM/Late Glacial time depth, characterized by an overall coalescence time estimate of 21–31 kya, and it probably originated in the southern region of Iran and split early into three branches. The first sub-clade, R2a, dates to ∼15–20 kya and includes several branches found mostly in southern Arabians (R2a1, R2a2, R2a3) and Persians (R2a4), as well as single mtDNAs from South Asia, the Near East and Europe. Two other mtDNA lineages, which are named here as R2b and R2c, restricted to Qashqais from southern Iran and potentially could have split before the main R2a sub-clade (sample size does not allow dating).  Figure 3. Maximum-parsimony phylogenetic tree of complete mtDNA sequences belonging to haplogroup R2. Overall, the complete mtDNA sequence analysis revealed an extremely high level of genetic diversity in the Iranian populations studied which is comparable to the other groups from the South Caucasus, Anatolia and Europe. The results of AMOVA and MDS analyses did not associate any regional and/or linguistic group of populations in the Anatolia/Caucasus and Iran region pointing to strong genetic affinity of Indo-European speaking Persians and Turkic-speaking Qashqais, thus suggesting their origin from a common maternal ancestral gene pool. The pronounced influence of the South Caucasus populations on the maternal diversity of Iranian Azeris is also evident from the MDS analysis results. Our results confirms that populations from Iran, Anatolia, the Caucasus and the Arabian Peninsula display a common set of maternal lineages although considerable regional differences in haplogroup frequencies exist [11], [29]. Meanwhile, some haplogroups previously defined as South Asian (such as R2 and HV2) could be considered as having Southwest Asian origin, taking into account the relatively high frequency and diversity of those haplogroups in Iran. Although R2 is a very rare haplogroup, the phylogeographic analysis indicates that it is present mostly in southern Arabia, while it has been suggested that the unrepresented Near East can be considered as a possible place of origin for R2 [37]. Meanwhile, our data indicate that haplogroup R2 has a likely pre-LGM/LGM time depth, with a coalescence time estimate of 21–31 kya, and it probably originated in southern Iran, although the neighboring Gulf Oasis region cannot be excluded, taking into account the close genetic affinity between Persians and Arabians proposed by Terreros et al. [26]. One should note also that the age estimate for the extremely rare haplogroup N3, which is specific for Iranian populations, is close to the LGM time, as it has been dated to 13–24 kya. dx.doi.org/10.1371/journal.pone.0080673 |
|
|
|
Post by Admin on May 2, 2016 22:47:26 GMT
 Genetic evidence indicates that most of the ethno-linguistic groups in India descend from a mixture of two divergent ancestral populations: Ancestral North Indians (ANI) related to West Eurasians (people of Central Asia, the Middle East, the Caucasus, and Europe) and Ancestral South Indians (ASI) related (distantly) to indigenous Andaman Islanders.1 The evidence for mixture was initially documented based on analysis of Y chromosomes2 and mitochondrial DNA3, 4, 5 and then confirmed and extended through whole-genome studies.6, 7, 8 Archaeological and linguistic studies provide support for the genetic findings of a mixture of at least two very distinct populations in the history of the Indian subcontinent. The earliest archaeological evidence for agriculture in the region dates to 8,000–9,000 years before present (BP) (Mehrgarh in present-day Pakistan) and involved wheat and barley derived from crops originally domesticated in West Asia.9, 10 The earliest evidence for agriculture in the south dates to much later, around 4,600 years BP, and has no clear affinities to West Eurasian agriculture (it was dominated by native pulses such as mungbean and horsegram, as well as indigenous millets11). Linguistic analyses also support a history of contacts between divergent populations in India, including at least one with West Eurasian affinities. Indo-European languages including Sanskrit and Hindi (primarily spoken in northern India) are part of a larger language family that includes the great majority of European languages. In contrast, Dravidian languages including Tamil and Telugu (primarily spoken in southern India) are not closely related to languages outside of South Asia. Evidence for long-term contact between speakers of these two language groups in India is evident from the fact that there are Dravidian loan words (borrowed vocabulary) in the earliest Hindu text (the Rig Veda, written in archaic Sanskrit) that are not found in Indo-European languages outside the Indian subcontinent.12, 13  Figure 1 Principal Component Analysis Admixture Dates To date ANI-ASI mixture, we capitalized on the fact that admixture between two populations generates allelic association (linkage disequilibrium [LD]) between pairs of SNPs.34 The LD decays at a constant rate as recombination breaks down the contiguous chromosomal blocks inherited from the ancestral mixing populations. The expected value of the admixture LD is related to the genetic distance between SNPs (the probability of recombination per generation between them) and the time that has elapsed since mixture.34 We previously reported simulations showing that dating population mixture based on the scale of admixture LD is robust to the use of imperfect surrogates for the ancestral populations, fine-scale errors in the genetic map, and a history of founder events in the admixed population, and is able to provide unbiased estimates for the dates of events up to 500 generations ago.26, 28, 29 We confirmed this by using new simulations with demographic parameters relevant to India (Appendix A). We estimated admixture dates for all the groups on the Indian cline with more than five samples (a minimum sample size is important for measuring LD with precision). We observe a decay of LD with genetic distance for all groups (Figures 2 and S3). By fitting an exponential function using least-squares (via rolloff), our point estimates for the dates range from 64 to 144 generations ago, or 1,856 to 4,176 years assuming 29 years per generation.35 We highlight two implications of these dates. First, nearly all groups experienced major mixture in the last few thousand years, including tribal groups like the Bhil, Chamar, and Kallar that might be expected to be more isolated. Second, the date estimates are typically more recent in Indo-Europeans (average of 72 generations) compared to Dravidians (108 generations). A jackknife estimate of the difference is highly significant at 35 ± 8 generations (Z = 4.5 standard errors from zero) (Table 1). A possible explanation is a secondary wave of mixture in the history of many Indo-European groups, which would decrease the estimated admixture date.  Focusing on the largest set of Indo-Europeans (four groups) and the largest set of Dravidians (five groups) consistent with mixture of the same ANI and ASI ancestral populations, we find that the expected and observed admixture LD amplitudes are equivalent to within the limits of our resolution. We restricted this analysis to Indian groups genotyped on Affymetrix arrays because this allowed us to analyze about 2.5 times more SNPs (n = 210,482), which improves the accuracy of inferences based on admixture LD. Limiting our analysis to samples genotyped on Affymetrix arrays raised the challenge that we could not use Georgians as part of our admixture graph fitting (we need a second West Eurasian outgroup to obtain tight constraints on the absolute estimates of ANI-ASI admixture), but in Appendix D we show that we can accurately infer the difference between the two amplitude values (observed − expected) even without access to Georgians by constraining the admixture proportions estimated via f4 ratio estimation. For both the Indo-European and Dravidian rank 1 sets, the observed amplitudes are statistically consistent with the expected values (Table 3). Thus, our data are consistent with all of the ANI ancestry in some selected sets of Indians (including groups speaking both Indo-European and Dravidian languages) being due to admixture events that we can date to within the past few thousand years. Accounting for statistical uncertainty, we estimate that the ANI ancestry that cannot be explained by a single wave of admixture in the last few thousand years has a 95% confidence interval (truncated to 0) of 0%–19% for Indo-Europeans and 0%–16% for Dravidians. Thus, all the ANI ancestry in some groups is consistent with deriving from admixture events that have occurred in the past few thousand years. Our analysis documents major mixture between populations in India that occurred 1,900–4,200 years BP, well after the establishment of agriculture in the subcontinent. We have further shown that groups with unmixed ANI and ASI ancestry were plausibly living in India until this time. This contrasts with the situation today in which all groups in mainland India are admixed. These results are striking in light of the endogamy that has characterized many groups in India since the time of mixture. For example, genetic analysis suggests that the Vysya from Andhra Pradesh have experienced negligible gene flow from neighboring groups in India for an estimated 3,000 years.1 Thus, India experienced a demographic transformation during this time, shifting from a region where major mixture between groups was common and affected even isolated tribes such as the Palliyar and Bhil to a region in which mixture was rare.  Figure 2 Dates of Mixture Our estimated dates of mixture correlate to geography and language, with northern groups that speak Indo-European languages having significantly younger admixture dates than southern groups that speak Dravidian languages. This shows that at least some of the history of population mixture in India is related to the spread of languages in the subcontinent. One possible explanation for the generally younger dates in northern Indians is that after an original mixture event of ANI and ASI that contributed to all present-day Indians, some northern groups received additional gene flow from groups with high proportions of West Eurasian ancestry, bringing down their average mixture date. This hypothesis would also explain the nonexponential decays of LD in many northern groups and their higher proportions of ANI ancestry. A prediction of this model is that some northern Indians will have genomes consisting of long stretches of ANI ancestry interspersed with stretches that are mosaics of both ANI and ASI ancestry (inherited from the initial mixture). Although we have not been able to test the predictions of this hypothesis, it may become possible to do so in future by developing a method to infer the ancestry at each locus in the genome of Indians that can provide accurate estimates even in the absence of data from ancestral populations. The dates we report have significant implications for Indian history in the sense that they document a period of demographic and cultural change in which mixture between highly differentiated populations became pervasive before it eventually became uncommon. The period of around 1,900–4,200 years BP was a time of profound change in India, characterized by the deurbanization of the Indus civilization,39 increasing population density in the central and downstream portions of the Gangetic system,40 shifts in burial practices,41 and the likely first appearance of Indo-European languages and Vedic religion in the subcontinent.18, 19 The shift from widespread mixture to strict endogamy that we document is mirrored in ancient Indian texts. The Rig Veda, the oldest text in India, has sections that are believed to have been composed at different times. The older parts do not mention the caste system at all, and in fact suggest that there was substantial social movement across groups as reflected in the acceptance of people with non-Indo-European names as kings (or chieftains) and poets.42 The four-class (varna) system, comprised of Brahmanas, Ksatriyas, Vaisyas, and Sudras, is mentioned only in the part of the Rig Veda that was likely to have been composed later (the appendix: book 10).42 The caste (jati) system of endogamous groups having specific social or occupational roles is not mentioned in the Rig Veda at all and is referred to only in texts composed centuries after the Rig Veda, for example, the law code of Manu that forbade intermarriage between castes.43 Thus, the evolution of Indian texts during this period provides confirmatory support as well as context for our genetic findings. DOI: dx.doi.org/10.1016/j.ajhg.2013.07.006 |
|
|
|
Post by Admin on Jul 24, 2016 21:44:49 GMT
 India has been underrepresented in genome-wide surveys of human variation. We analyse 25 diverse groups in India to provide strong evidence for two ancient populations, genetically divergent, that are ancestral to most Indians today. One, the ‘Ancestral North Indians’ (ANI), is genetically close to Middle Easterners, Central Asians, and Europeans, whereas the other, the ‘Ancestral South Indians’ (ASI), is as distinct from ANI and East Asians as they are from each other. By introducing methods that can estimate ancestry without accurate ancestral populations, we show that ANI ancestry ranges from 39-71% in most Indian groups, and is higher in traditionally upper caste and Indo-European speakers. Groups with only ASI ancestry may no longer exist in mainland India. However, the indigenous Andaman Islanders are unique in being ASI-related groups without ANI ancestry. Allele frequency differences between groups in India are larger than in Europe, reflecting strong founder effects whose signatures have been maintained for thousands of years owing to endogamy. We therefore predict that there will be an excess of recessive diseases in India, which should be possible to screen and map genetically. We warn that ‘models’ in population genetics should be treated with caution. Although they provide an important framework for testing historical hypotheses, they are oversimplifications. For example, the true ancestral populations of India were probably not homogeneous as we assume in our model, but instead were probably formed by clusters of related groups that mixed at different times. However, modelling them as homogeneous fits the data and seems to capture meaningful features of history.  The following figure illustrates the general model which looms in the background of this paper: Note that the Andaman Islanders, the Onge, aren’t really the ancestors of Indians on the mainland. Rather, they’re a branch of the ancient population which presumably first settled South Asia, and close to the ASI. Who were the ASI? Since they aren’t really around, we can only generate conjectures and inferences. In this paper the ANI are actually represented in some ways by Europeans, even though presumably the assumption is that both these are daughter populations of another group. Though not pushed very hard, they do mention proto-Indo-Europeans as the candidate for the ANI.  This should not surprise, previous work shows that South Asians distribute along an axis away from Europeans. One of the points in the paper is that there is both geographic and caste stratification. I added some labels, but I thought drilling-down was probably useful. I don’t know all these groups off the top of my head, and I assume few of readers do either. So I zoomed in:  I think some of the shortcomings with a sample size on the order of the low hundreds is rather clear. They couldn’t even use all their samples, or some of the samples were not relevant to the question on hand. The Siddis are an Indian-African mix which emerged during the period of Muslim domination when that group imported black slaves. The Tibeto-Burman groups of Northeast India are interesting, but outliers. The general trends are clear, North Indian groups have more ANI than South Indian groups, and upper caste groups have more ANI than lower caste groups, but that is only with “all things equal.” Note that upper caste South Indian groups clearly have more ANI than lower caste South Indians, but they have a lower proportion than some North Indian lower castes, and are in the range of one North Indian tribal group. Some of the outliers are also interesting; the lower caste individual similar to Austro-Asiatic tribals is from a group which resides in a region with many Austro-Asiatic peoples. Clearly there has been identity switching, so you have aberrations such as one North Indian tribal who clusters with Kashmiri Pandit Brahmins! The Austro-Asiatic group is also interesting, because they speak languages related to those of Southeast Asia. Here is a map of the Austro-Asiatic languages:  We know with near 100% certainty that much of Burma & Thailand were dominated by Mon-Khmer languages before the arrival of the Shan, Bamar (Burmans) and Thai peoples (to mention a few). This is matter of historical record, the rise of modern Burma and Thailand was largely a story of the eclipse of Mon and Khmer societies who transmitted to them much of the Indic character which they have (e.g., the northern populations often arrived as Mahayana Buddhists, but the Mon and Khmer Theravada Buddhism was adopted as the dominant religions in the new states). The position of the Munda languages is more confused, as some posit that they arrived from the east, while others argue that the the Austro-Asiatic languages expanded east from India. This is not going to be resolved in this blog post, but let me note that the genetic data above, which show an “eastern” affinity of the Munda, can be combined to with cultural datum such as the arrival of rice farming from the east and historical records which document the migration of populations from Burma, to construct a plausible east-west narrative. In contrast it seems an almost default position by many that the Austro-Asiatics are the most ancient South Asians, marginalized by Dravidians, and later Indo-Europeans. I would not be surprised if it was actually first Dravidians, then Austro-Asiatics and finally Indo-Europeans. Dravidian are found in every corner of the subcontinent (Brahui in Pakistan, a few groups in Bengal, and scattered through the center) while the Austro-Asiatics exhibit a more restricted northeastern range.  Interestingly, one of the GIH subgroups fall outside the main gradient of Indian groups, suggesting that they harbor substantial ancestry that is not a simple mixture of ASI and ANI. A speculative hypothesisis that some Gujarati groups descend from the founders of the “Gurjara Pratihara” empire, which is thought to have been founded by Central Asian invaders in the 7th century A.D. and to have ruled parts of northwest India from the 7-12th centuries. I. Karve noted that endogamous groups with names like “Gurjar” are now distributed throughout the northwest of the subcontinent, and hypothesized that that they likely trace their names to this invading group.  Next are two charts which shows Indians, Europeans, and Chinese. In the first the PCA was originally constructed with Europeans & Chinese, and the Indians were projected onto it using the variation found in the first two groups. In the second case, Indians and Chinese were used to construct the PCA, and Europeans projected.  What you see is that Europeans are all equally related to Indians, but Indians exhibit a gradient of relationship to Europeans. That is, there is no European group which in particular resembles Indians via the connection with ANI; the distance between all European groups and ANI seems roughly equal. The Indians vary in their relationship to Europeans because they vary in their proportion of ANI. In the table above there is a reference to the proportion of ANI and ASI in each Indian group. One question you might ask: how do you estimate the proportions of ancestry from groups which you don’t have any information about because they no longer exist? Europeans and the Onge can serve as proxies for the ANI and ASI respectively, but how far does this get you? Well, the methods that they used (they have three) which determine ancestral proportions can be used on populations which exist. So here is a figure which shows how their methods compare when you look at a population where we know something concrete about their ancestral populations because those ancestral populations are still extant, African Americans:  We propose that the high FST among Indian groups could be explained if many groups were founded by a few individuals, followed by limited gene flow. This hypothesis predicts that within groups, pairs of individuals will tend to have substantial stretches of the genome in which they share at least one allele at each SNP. We find signals of excess allele sharing in many groups. Six Indo-European- and Dravidian speaking groups have evidence of founder events dating tomore than 30 generations ago…including the Vysya at more than 100 generations ago…Strong endogamy must have applied since then (average gene flow less than 1 in 30 per generation) to prevent the genetic signatures of founder events from being erased by gene flow. Some historians have argued that ‘caste’ in modern India is an ‘invention’ of colonialism in the sense that it became more rigid under colonial rule. However, our results indicate thatmany current distinctions among groups are ancient and that strong endogamy must have shaped marriage patterns in India for thousands of years. Two features of the inferred history are of special interest. First, the ANI and CEU form a clade, and further analysis shows that the Adygei, a Caucasian group, are an outgroup. Many Indian and European groups speak Indo-European languages, whereas the Adygei speak a Northwest Caucasian language. It is tempting to assume that the population ancestral to ANI and CEU spoke ‘Proto-Indo-European’, which has been reconstructed as ancestral to both Sanskrit and European languages, although we cannot be certain without a date for ANI-ASI mixture. Reich D, Thangaraj K, Patterson N, Price AL, Singh L. 2009. Reconstructing Indian population history. Nature 461:489-494. doi:10.1038/nature08365 |
|
|
|
Post by Admin on Aug 19, 2016 21:18:33 GMT
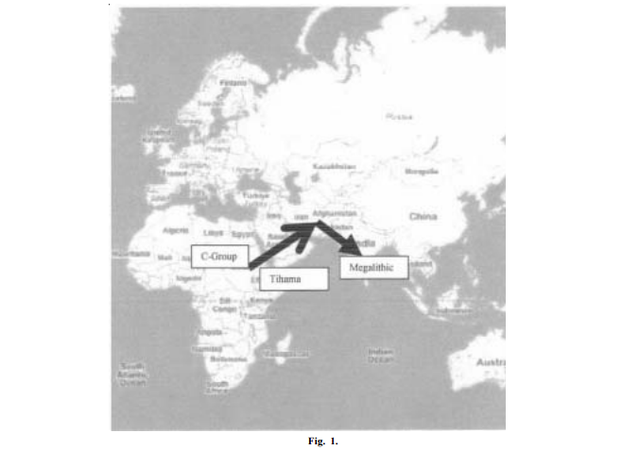 Researchers have long speculated on the possibility of Dravidian speakers migrating through Iran to India. This view is supported by the presence of Indian haplogroups in Iran (Gonzalez et al. 2007), and the close relationship between the Dravidian and Elamite languages (McAlpin 1974, 1981). Eleven of Iran’s M haplogroups are found in India. In Iran hg M is found predominately in the Sussa region. Around 5% of Iranians carry the M haplogroup. The most frequent Indian haplogoup in Iran is M3 (Metspula et al. 2004). Even though most molecular anthropologists believe the Dravidians originated in situ in India.  The spread of common archaeological assemblages associated with the C-Group, genetically related languages and genes from Africa across Arabia and Iran into India support a recent expansion of Dravidian speaking people from Africa to India. The archaeological and molecular evidence provides footprints of a recent hg M ancestral migration from Nubia to India. The existence of the L3a-M motif in the Senegambia characterized by the DdeI site np 10394 and AluI site np 10397 in haplotype AF24; the presence of the nucleotides characteristic of the Indian macrohaplogroup M in Africa and Arabia; and the reality that M1 does not descend from an Asian M macrohaplogroup (Sun et al. 2005) make a ‘back migration’ of M1 to Africa highly unlikely.  The geographical distribution of the archaeological signature of the C-Group people from Nubia to India matches the location of populations carrying hg M. The presence of Indian M sequences in Africa, Arabia, Iran and Yemen (Gonzalez et al.. 2006) in conjunction with the linguistic (Aravanan 1976, 1979; Upadhyaya and Upadhyaya 1976, 1979), archaeological (Lal 1963; Lahovary 1963; Rao 1972) and anthropological (Nayar 1977; Sergent 1995; Sastri 1966) evidences suggest that the Dravidian speakers formerly lived in Nubia and migrated to India over 5000 years ago and the Indian M macrohaplogroups do not have an in situ origin. Int J Hum Genet, 8(4): 325-329 (2008) |
|























