|
|
Post by Admin on Aug 6, 2017 19:08:47 GMT
 Figure 5 QQ plot showing observed versus expected −log10 (p values) for association at all Neanderthal-derived loci of the shared variance of all Neanderthal-associated brain measures. We further examined this genetic region to assess the plausibility of its Neanderthal introgression. The 53 kb block we identified overlaps a previously predicted Neanderthal-introgressed region – by both Vernot et al.30 and Sankararaman et al.5. Since regions of extended similarity to archaic genomes could arise by incomplete lineage sorting (ILS), we calculated the maximum possible length of such an ILS region given the recombination rate and time since introgression31. Assuming the lowest average recombination rate in the region (which is most conservative), the branch lengths in years8, a generation time of 25 years, and a mutation rate of either 0.5 × 10−9 per site per year or 1 × 10−9 per site per year, we can rule out that any region longer than 1580 bp (0.5 × 10−9) or 3163 bp (1 × 10−9) is the result of ILS. The chance of maintaining a 53 kb block in both lineages is conservatively estimated at p < 9.4 × 10−7, using a gamma distribution with shape parameter 2 and a conservative rate parameter of 1/3163 bp31. Discussion In this work, we describe relationships between Neanderthal-derived genetic variation and co-localized cranial and brain morphology in modern humans. The results show that greater NeanderScore is associated with more Neanderthal-like skull shape (corresponding to published shape differences between modern humans and Neanderthals as documented in fossil samples2 and shown in Fig. 1), as well as regional changes in brain morphology underlying these skull changes, specifically in the IPS and visual cortex. This work not only offers an unprecedented window into structure of the Neanderthal brain, but also characterizes the contributions of admixture with H. neanderthalensis to the evolution of the modern human brain. In examining the associations of NeanderScore with skull shape, it is important to note that the topology of the identified occipito-parieto-temporal patch associated with NeanderScore specifically recapitulates the pattern of expansion in Neanderthal relative to anatomically modern human skulls previously reported from fossil remains (Fig. 1, right)2. This finding provides crucial validation of the NeanderScore measurement and suggests that even in the context of the modern H. sapiens genome, Neanderthal genetic variation is associated with patterns of skull dimensions that mirror known Neanderthal phenotypes.  Figure 6 Derivation of NeanderScore. Phylogenic tree showing relationship of Yoruba, a genotyped individual, Neanderthal and Primates. At a given SNP, Yoruba and Primates contain the ancestral allele (A), Neanderthal contains the derived allele (B) and the genotyped individual may have either allele (A,B). Additional validation of the NeanderScore metric lies in its comparison with other reports characterizing Neanderthal-derived genetic contributions to modern humans. The coefficient of variation (i.e., the relative standard deviation, independent of the mean and comparable across measures) of the NeanderScore metric is consistent with previous published studies5 that were performed with a different goal: prior studies, unlike ours, sought to test for differences in admixture across populations, and have, therefore, calculated the proportion of the entire genome that was derived from Neanderthals. In those studies, approximately 1.15% of the entire genome of persons of European decent was found to be derived from Neanderthals (standard deviation = 0.08)5. Inherently, that approach and the present one capture related though different characteristics of Neanderthal-derived genetic information and the means (and therefore the standard deviations as well) of these differently derived measures are not the same. Importantly, however, the coefficient of variation in previous studies was also 0.070,5 identical to the coefficient of variation reported here, providing further validity to the NeanderScore measure. In determining NeanderScore related changes in brain morphology, we found two significant cortical regions, the IPS and primary visual cortex, which both directly underlie the region of skull shape associations. The IPS, though present throughout highly gyrified modern primates28, has been theorized to have undergone substantial evolutionary expansion in hominids, with cross-species functional neuroimaging demonstrating unique visuospatial processing characteristics in modern humans relative to rhesus monkeys32. Additionally, cranial vault analyses of fossil skulls have suggested differences in the parietal lobes of Neanderthals14, 15, and the intraparietal sulcal region in particular has been hypothesized to be a focus for some of these differences33. Moreover, the fact that the IPS is particularly critical for tool manipulation in modern humans34 makes this finding even more intriguing in view of continued debate over the nature and development of Neanderthal tool use35. |
|
|
|
Post by Admin on Aug 7, 2017 19:00:47 GMT
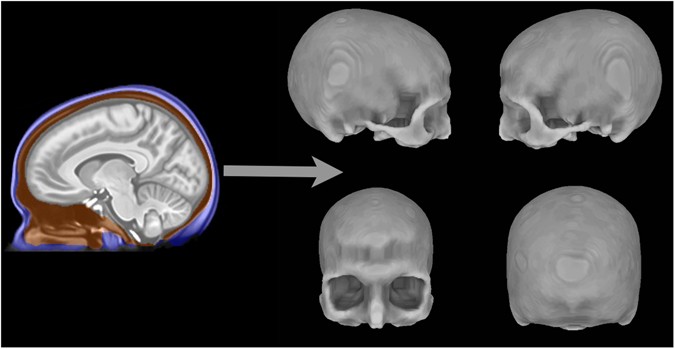 Figure 7 Skull surface creation from the T1-weighted MRI scan of a single participant. Left shows the segmentation procedure performed in the volume where voxels labeled blue represent scalp, orange voxels represent skull, and gray voxels represent the intracranial area. Right shows the 3D skull surface derived from voxels labeled as skull (orange) in the segmentation. The other brain region revealed to have significant associations with NeanderScore was primary visual cortex. This cortical region is responsible for the first steps in processing of visual information in the mammalian cortex and feeds into later brain regions in the ventral and dorsal visual processing streams (which differentially subserve object recognition and visuospatial object location, respectively)36, with the IPS playing a prominent role in the latter. Though the functioning of the primary visual system is relatively conserved in primates, the size of primary visual cortex in modern humans is smaller than would be expected from brain volume37. Our data not only suggest that this may be less the case in Neanderthals, but also are consistent with cranial remains showing more prominent visual systems in Neanderthals than in modern humans15.  It should be noted that we did not find associations of NeanderScore with smaller frontotemporal volumes38 or shortened anterior extension of the temporal lobes13, as might have been hypothesized from previous cranial analyses of H. neanderthalensis, suggesting either that these particular phenotypes, if accurate, are not driven by the allelic variation captured by the NeanderScore measurement, or that such effects are more directly influenced by any of the myriad factors that establish the genetic and biological context of this modern cohort. Additionally, some of these phenotypes may be only partially modulated by genotypic factors inherited from Neanderthals, with an effect too small to be observed in our sample. In contrast, the effect sizes we observed here for the associations of NeanderScore with the skull and brain measures were moderate (average Cohen’s d = 0.58) but appropriate for the sample sizes used. The analyses reported here were restricted to a sample of individuals of European descent. It is known that the degree of admixture is variable in different modern populations. For example, East Asian populations have been found to have a larger portion of the genome derived from Neanderthals than European populations (up to 20% more), though the admixed regions of the genome are not necessarily overlapping5. This raises the possibility that the findings reported here may not translate to other populations, where Neanderthal introgression may involve other genomic regions that may be functional in different ways. As large neuroimaging and genetic data from different populations become available, future work could investigate this possibility by performing similar analyses in different populations, including using African populations with minimal Neanderthal admixture as potential “null hypothesis” groups.  Finally, our analyses of the relationship of our findings to specific Neanderthal-derived gene variants, revealed a single 53 kb LD block that was significantly associated with the shared variance of the identified Neanderthal-derived brain and skull changes and that encodes for the gene GPR26. In line with our primary hypothesis for this analysis, that such genetic influences would be found in genes preferentially expressed in the human brain, GPR26 in fact encodes a G-protein coupled receptor subtype that is preferentially expressed in the brain29. Interestingly, in human post-mortem brains samples, expression of GPR26 peaks perinatally39, when the visual system is first challenged, indicating that it may play a role in development of the human visual system. Mouse models also show this gene to impact both affective and energy homeostatic functions40, 41. Additionally, this G-protein-coupled receptor has been shown to form oligomers with the 5-HT1a receptor42, perhaps providing a putative mechanism underlying the Neanderthal-related brain changes found here. Although the nature of the influence of this region on modern and archaic human nervous systems is uncertain, a possible link between brain energy regulation, neurodevelopment and mature structure may merit further investigation. Because the NeanderScore measure employed here is, itself, polygenic, it is likely that the genetic contributions to skull and brain morphology we observed involve a number of different genetic loci. Nonetheless, our exploratory post-hoc genome-wide analysis of the shared variance of these findings identified only a single significant region. It is unlikely that this single locus, the LD block on chromosome 10, fully explains the brain and skull findings, and in fact, the Manhattan plot in Fig. 4 suggests that multiple other regions that do not meet strict Bonferroni corrections may represent some degree of true signal. Our modest sample size may have been underpowered to identify these additional signals. As larger datasets containing both genotype and neuroimaging data of brain and skull become available, future work will likely uncover additional genetic regions contributing to these findings. Taken together, the associations between Neanderthal sequence variation and co-localized skull and brain morphology in modern humans engender an enduring, living footprint of H. neanderthalensis – a residual echo of shared, intimate history with a fallen lineage close to our own. To the extent that characterization of Neanderthal variation in present-day people can provide insights into archaic human phenotypes, this work can form the basis of future studies aimed at a more thorough understanding of Neanderthal biology. By the same token, we suggest that Neanderthal gene flow into modern humans is not only of evolutionary interest, but may also be functional in the living H. sapiens brain, revealing novel genetic influences on neurodevelopment of the visuospatial system upon which a fuller account of molecular mechanisms of IPS-driven normative mental functions, such as visuospatial integration and tool manipulation, can be built. This, in turn, may inform models of IPS-associated cognitive disability as seen in select developmental and neurological disorders43,44,45,46. Scientific Reports 7, Article number: 6308 (2017) |
|
|
|
Post by Admin on Sept 27, 2017 19:13:13 GMT
 Figure 1 Schematic Representation of Rind and Rpop Recent analyses have found that a substantial amount of the Neandertal genome persists in the genomes of contemporary non-African individuals. East Asians have, on average, higher levels of Neandertal ancestry than do Europeans, which might be due to differences in the efficiency of purifying selection, an additional pulse of introgression into East Asians, or other unexplored scenarios. To better define the scope of plausible models of archaic admixture between Neandertals and anatomically modern humans, we analyzed patterns of introgressed sequence in whole-genome data of 379 Europeans and 286 East Asians. We found that inferences of demographic history restricted to neutrally evolving genomic regions allowed a simple one-pulse model to be robustly rejected, suggesting that differences in selection cannot explain the differences in Neandertal ancestry. We show that two additional demographic models, involving either a second pulse of Neandertal gene flow into the ancestors of East Asians or a dilution of Neandertal lineages in Europeans by admixture with an unknown ancestral population, are consistent with the data. Thus, the history of admixture between modern humans and Neandertals is most likely more complex than previously thought. As modern humans migrated out of Africa and dispersed throughout the world, they encountered and hybridized with Neandertals.1, 2 The similarly low levels of Neandertal ancestry found in all modern non-African populations studied to date have been parsimoniously interpreted to be the result of a single pulse of admixture into the population ancestral to all non-Africans. However, recent reports show that East Asians have, on average, inherited ∼20% more Neandertal ancestry than Europeans have.3, 4, 5, 6 Two explanations have been proposed to account for this observation. Sankararaman et al.5 suggested that because Neandertal lineages appear to be subject to widespread purifying selection in modern humans, differences in the efficiency of purifying selection could account for higher levels of Neandertal ancestry in East Asians. This hypothesis is supported by previous studies that have shown that East Asians have a smaller effective population size than do Europeans.7 In contrast, through extensive simulations, Vernot and Akey4 found that the excess of Neandertal ancestry in East Asians could not be explained by a single ancestral introgression event. Rather, the data were better explained by a two-pulse model, where introgression occurred in the common ancestor of East Asians and Europeans and was followed by additional gene flow into East Asians. However, these simulations did not account for the potential confounding effects of natural selection, and thus ambiguity remains about whether a simple single pulse of admixture between modern humans and Neandertals can explain the data. To investigate how patterns of introgressed Neandertal sequences in East Asians and Europeans are influenced by potential differences in the efficiency of purifying selection between populations, we first partitioned the genome by using B-values,8 which measure the degree to which neutral variation has been reduced as a result of linked selected sites. Specifically, we selected 50-kb windows from Vernot and Akey4 and binned each window according to the minimum B-value at any base in that window. B-values range from 0 (all neutral diversity eliminated by background selection) to 1 (no reduction of neutral diversity by background selection). Next, we calculated values of two summary statistics of Neandertal introgression,4 Rind and Rpop (Figure 1), as a function of B-values. Rind is the ratio of the amount of introgressed Neandertal sequence per individual in East Asians to that in Europeans (Figure 1). Rpop is the ratio of the number of genomic bases covered by introgressed Neandertal sequence in any East Asian individual to that in any European individual (we corrected for differences in sample size by subsampling sets of 20 individuals from each population; Figure 1). Our previously measured genome-wide values of Rind and Rpop were 1.21 and 1.05, respectively.4 Moreover, Rind has consistently been reported to be greater than 1, including values of 1.19,3 1.20,5 and 1.4.6 These estimates indicate that although approximately the same amount of the Neandertal genome survives in Europeans and East Asians, a given Neandertal haplotype in East Asians is on average at higher frequency (Figure 1).  Figure 2 Estimates of Rind and Rpop as a Function of B-Value Cutoffs If elevated levels of Neandertal ancestry in East Asians are due to differences in the efficiency of purifying selection between East Asians and Europeans, then Rind should vary significantly by B-value. Genomic regions under strong purifying selection would be more strongly depleted in Neandertal ancestry in the historically larger European population, leading to high Rind at low B-values and Rind closer to 1 at high B-values. In contrast, we observed that Rind was fairly stable with increasing B-value cutoffs (Figure 2). For example, the estimate of Rind in regions with a minimum B-value of 0.975 (spanning ∼106 Mb of the genome), which indicates that neutral variation was reduced by <2.5% as a result of background selection, was 1.175. Thus, in this more neutral subset of the genome, East Asian individuals have on average 17.5% more introgressed sequence than Europeans. This percentage closely parallels genome-wide estimates. Unlike Rind, which remained stable with increasing B-values, Rpop showed a marked decline as the B-value cutoff exceeded 0.850, indicating that in more neutrally evolving genomic regions, less of the Neandertal genome survives at the population level in East Asians than in Europeans. This observation is consistent with previous studies, which have found that a smaller ancestral effective population size in East Asians than in Europeans results in more intense genetic drift.7 Higher genetic drift would result in the loss of low-frequency Neandertal haplotypes; this effect would be strongest in regions where the competing force of purifying selection is weakest.9 Qualitatively, patterns of Rind and Rpop as a function of B-value suggest that the excess of Neandertal ancestry in East Asians cannot be explained by differences in selective forces alone and that a model of a single ancestral pulse of Neandertal introgression is unlikely. To more formally evaluate demographic models compatible with patterns of Neandertal ancestry in East Asians and Europeans, we performed approximate Bayesian computation (ABC)10, 11 on putatively neutral sequence by calculating Rind and Rpop in genomic regions with a minimum B-value ≥ 0.975. ABC analysis involves simulating data under different demographic models, calculating summary statistics from these simulations, and selecting simulations in a principled manner that best matches the observed summary statistics. We first simulated neutral sequence data under (1) a one-pulse model where all Neandertal sequence introgressed in a single pulse into the common ancestor of East Asians and Europeans (m1; Figure 3A) and (2) a two-pulse model with varying amounts of additional introgression into either Europeans or East Asians (m2; Figure 3A) after population splitting. Specifically, we performed simulations in which m2 / m1 ranged from −2% to 33% (negative values indicate simulations with additional introgression into Europeans; m2 = 0 is a one-pulse model). For summary statistics, we used both Rind and Rpop. We have previously shown that these statistics can distinguish between archaic admixture demographic models.4 We performed >30,000 simulations and estimated demographic parameters by using ABC with the R package “abc.”13 Specifically, we used the “abc” function with a non-linear neural-network regression method of correcting accepted parameter values.10 Results were similar when we corrected values by local linear regression. A complete description of the simulated demographic models can be found in Appendix A. |
|
|
|
Post by Admin on Sept 29, 2017 18:53:58 GMT
 Figure 3 Inference of Admixture Models from Genomic Regions with Little or No Selective Constraint Only simulations with additional introgression into East Asians (two-pulse models) were accepted as plausible in the ABC analysis (Figure 3A). We estimated that a second pulse of 15% more introgression into East Asians could explain the observed excess of Neandertal introgression (95% confidence interval [CI] of m2 / m1 = 6.8%–26.6%). Under the null hypothesis of a single pulse of admixture, the ratio m2 / m1 = 0%, which is well outside the preferred range. Given these results, the two-pulse model is significantly favored, and we can strongly reject the null hypothesis of a one-pulse model (p < 6.7 × 10−4). For completeness, we repeated our ABC analysis by using summary statistics from regions with a minimum B-value ≥ 0.950 (spanning 326 Mb of the genome), which again significantly favored the two-pulse model (Figure S1). Therefore, a one-pulse model is rejected both for genome-wide calculations of Neandertal ancestry4 and when regions that might have been subjected to selective constraint are excluded. It is important to stress that although a two-pulse model of admixture explains the empirical data significantly better than the simple one-pulse model considered here, it does not necessarily mean that the two-pulse model is correct. To investigate additional plausible demographic models, we also considered a model in which a single ancestral pulse of introgression was followed by admixture between the European population and a third modern human population that had not interbred with Neandertals. In this scenario, the amount of Neandertal ancestry in Europeans is effectively diluted by admixture with a population not carrying Neandertal lineages. Such a population could be from Africa, where there is expected to have been no, or little, Neandertal ancestry.14 Alternatively, it could be an unknown “ghost” Eurasian population that was entirely absorbed into Europeans. To determine whether a European-dilution model could explain the data, we again performed ABC by using the summary statistics Rind and Rpop, calculated from neutral subsets of the genome (B-value ≥ 0.975) as described above. Specifically, we simulated sequence data under a one-pulse model where all Neandertal sequence introgressed in a single pulse into the common ancestor of East Asians and Europeans (m1; Figure 3B) and the ancestral European population then admixed with a third population denoted as X (Figure 3B). We varied the proportion of modern-day European ancestry derived from population X (AX) from 0% to 35%. We estimated that the observed patterns of Neandertal introgression are compatible with a European-dilution model if an average of 18.2% of modern European ancestry (AX) was contributed by this third population (95% CI of AX = 9.2%–27.6%). This is significantly larger than current estimates of African ancestry in Europeans (1%–3%12 and 2.3%;15 p = 5.7 × 10−4), suggesting that migration from Africa to Europe cannot explain the larger amount of Neandertal ancestry in East Asians than in Europeans. Tantalizingly, recent work suggests that modern Europeans might comprise admixture of three ancestral groups.16 However, each of these groups is estimated to contain ∼2% Neandertal ancestry16 and thus could not have diluted the amount of Neandertal ancestry in modern Europeans enough to account for the differences with East Asians. Thus, on the basis of current evidence, differential migration seems less likely to explain the data, increasing the likelihood of multiple-pulse models.  In addition to evaluating models of the interactions between modern humans and Neandertals, we used these analyses of Neandertal ancestry to elucidate other aspects of human demographic history. For example, in addition to estimating the parameters m2 / m1 and AX, we found that the ratio of ancestral effective population sizes between Europeans and East Asians, NeEUR / NeASN, had a significant effect on the fit of our models. Using the same ABC analyses, we estimated NeEUR / NeASN to be 1.93 (95% CI = 1.57–2.73) under a two-pulse model and estimated NeEUR / NeASN to be 1.59 (95% CI = 1.35–1.89) under the European-dilution model (Figure S1). Both of these estimates are consistent with previously accepted values.7, 15 In summary, by focusing on putatively neutral regions of the genome, we have shown that the observed patterns of Neandertal ancestry in Europeans and East Asians are not consistent with a simple one-pulse model of admixture. Thus, differences in the efficiency of purifying selection among populations are unlikely to account for higher levels of Neandertal ancestry in East Asians than in Europeans. We have shown that more complex and nuanced models are necessary to explain the data and have furthermore suggested two such models that are consistent with observed patterns of Neandertal introgression in Europeans and East Asians. Additionally, we have shown that studies of Neandertal ancestry can be informative about other aspects of human history. Combined with the analysis of ancient DNA of archaic and modern humans, additional studies in geographically diverse populations will help narrow the space of plausible demographic models. Such models will provide critical insights into hominin evolutionary history and the key parameters governing admixture dynamics between modern humans and Neandertals. AJHG Volume 96, Issue 3, p448–453, 5 March 2015 |
|
|
|
Post by Admin on Oct 1, 2017 18:55:53 GMT
 An international team of researchers has conducted a new test of Neanderthal remains found at Vindija Cave in Croatia and found them to be older than previous studies indicated. In their paper published in the Proceedings of the National Academy of Sciences, the team describes their dating technique and the possible implications of their findings. The Neanderthal remains were originally found in the cave approximately 40 years ago and have been tested for age several times. They have also been the subject of much speculation, as it was thought that the remains represented the last of the Neanderthals in that part of Europe and that they existed for a short period of time in close proximity to modern humans. Initial testing suggested the remains were approximately 28,000 to 29,000 years old. More recent tests have put them at 32,000 to 34,000 years old.  Both time frames coincide with the arrival of modern humans into the area, keeping alive the theory that the two groups mixed, both physically and socially. But now, using what is being described as a more accurate technique, the group with this new effort has found that the remains are older than thought. The new technique, called ZooMS involves radiocarbon dating hydroxyproline—an amino acid taken from collagen samples found in bone remains. The team also purified the collagen to remove contaminants. The researchers report that the new technique indicates that the remains—all four samples—were approximately 40,000 years old. This new finding puts the Neanderthal in the cave well before the arrival of modern humans, thus, there could not have been mixing of the two.  The researchers also studied other artifacts from the cave, including other animal bones, and found that the artifacts were a mixed bag, representing a timeline of thousands of years. The animal bones, they found, were from bears. This has led the team to conclude that the reason more modern artifacts were found with older artifacts is because of bears mixing them up. Previous dating of the Vi-207 and Vi-208 Neanderthal remains from Vindija Cave (Croatia) led to the suggestion that Neanderthals survived there as recently as 28,000–29,000 B.P. Subsequent dating yielded older dates, interpreted as ages of at least ∼32,500 B.P. We have redated these same specimens using an approach based on the extraction of the amino acid hydroxyproline, using preparative highperformance liquid chromatography (Prep-HPLC). This method is more efficient in eliminating modern contamination in the bone collagen. The revised dates are older than 40,000 B.P., suggesting the Vindija Neanderthals did not live more recently than others across Europe, and probably predate the arrival of anatomically modern humans in Eastern Europe. We applied zooarchaeology by mass spectrometry (ZooMS) to find additional hominin remains. We identified one bone that is Neanderthal, based on its mitochondrial DNA, and dated it directly to 46,200 ± 1,500 B.P. We also attempted to date six early Upper Paleolithic bone points from stratigraphic units G1, Fd/d+G1 and Fd/d, Fd. One bone artifact gave a date of 29,500 ± 400 B.P., while the remainder yielded no collagen. We additionally dated animal bone samples from units G1 and G1–G3. These dates suggest a co-occurrence of early Upper Paleolithic osseous artifacts, particularly split-based points, alongside the remains of Neanderthals is a result of postdepositional mixing, rather than an association between the two groups, although more work is required to show this definitively. |
|












