Post by Admin on Aug 27, 2021 2:21:06 GMT
The Toalean burial from Leang Panninge
The most distinctive archaeological assemblages associated with Holocene hunter-gatherers in Wallacea belong to the Toalean technocomplex (8–1.5 kya)3,4,27,28. Found only in a 10,000 km2 area of South Sulawesi3 (Fig. 1b), Toalean cultural assemblages are generally characterized by backed microliths and small stone projectiles (‘Maros points’)4 (Extended Data Fig. 1a–c). In 2015, excavations at Leang Panninge in the Mallawa district of Maros, South Sulawesi (Fig. 1b), uncovered the first relatively complete human burial from a secure Toalean context (Extended Data Figs. 1–5, Supplementary Information). The individual was interred in a flexed position29 in a rich aceramic Toalean stratum. Exposed at a depth of around 190 cm, the burial has an inferred age of 7.3–7.2 kyr cal BP obtained from 14C dating of a Canarium sp. seed (Extended Data Figs. 2, 3, Supplementary Table 1). Morphological characters indicate that this Toalean forager was a 17–18-year-old female with a broadly Australo-Melanesian affinity, although the morphology does not fall outside the range of recent Southeast Asian variation (Supplementary Information).
Genomic analysis
We extracted ancient DNA from bone powder obtained from the petrous portion of the temporal bone of the Leang Panninge individual. After library preparation, we used a DNA hybridization capture approach to enrich for approximately 3 million single-nucleotide polymorphisms (SNPs) across the human genome (1240K and archaic admixture capture panels30) as well as for the entire mitochondrial genome (mtDNA capture31). We retrieved 263,207 SNPs on the 1240K panel, 299,047 SNPs on the archaic admixture panel and the almost complete mtDNA sequence. Authenticity of the analysed ancient DNA was confirmed by short average fragment length, elevated damage patterns towards the molecule ends, and low autosomal and mtDNA contamination estimates (Supplementary Fig. 1). We confirmed that the individual was genetically of female sex. Analysis of the polymorphisms present in the reconstructed mtDNA sequence suggests a deeply divergent placement within mtDNA haplogroup M (Supplementary Table 17, Supplementary Fig. 2).
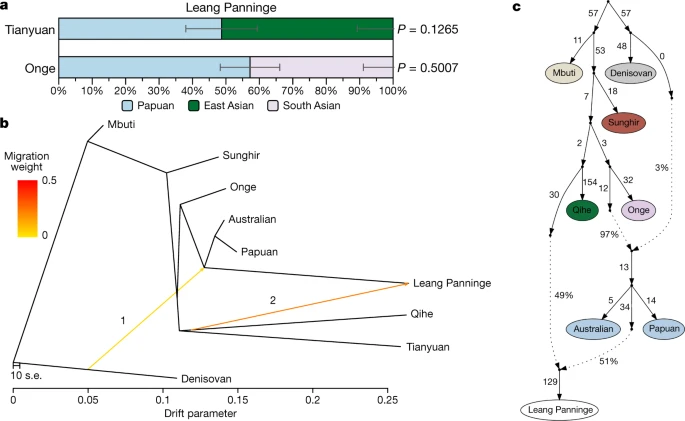
We initiated our genomic investigation by principal component analyses (PCAs), comparing the Leang Panninge genome with present-day individuals from East Asia, southeast Asia and Near Oceania (comprising Indigenous Australia, Papua New Guinea and Bougainville) genotyped on the Human Origins SNP panel18,32,33,34. The newly generated genome and relevant published genomes from ancient individuals from eastern Eurasia were then projected on the PCA1,34,35,36,37,38. Leang Panninge falls into PCA space not occupied by any present-day or ancient individuals, but is broadly located between Indigenous Australian peoples and the Onge (Fig. 2a, Extended Data Fig. 6). F3-statistics33 of the form f3 (Mbuti; Leang Panninge, X), where X is replaced with present-day Asian-Pacific groups, indicated that the new genome shares most genetic drift with Near Oceanian individuals (Fig. 2b). We confirmed these results with f4-statistics33, suggesting similar affinity of Leang Panninge and Papuan individuals to present-day Asian individuals, despite Near Oceanian groups forming a clade to the exclusion of Leang Panninge (Extended Data Fig. 7a, b). All present-day groups from the region, with the exception of the Mamanwa and the Lebbo26, carry only a minor contribution of Papuan-related ancestry (Supplementary Fig. 4).
Fig. 2: The Leang Panninge genome within the regional genetic context.
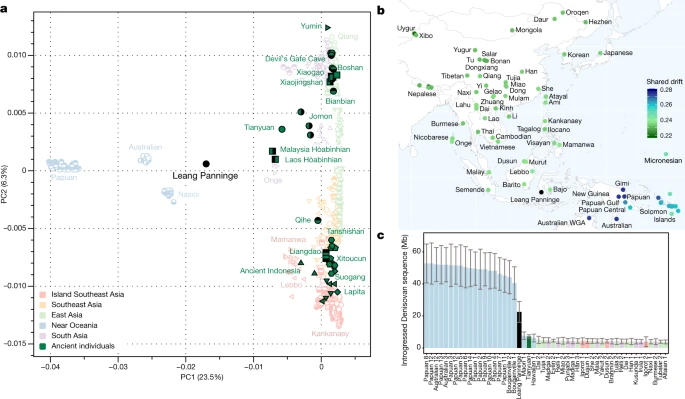
a, PCA calculated on present-day individuals from eastern Eurasia and Near Oceania, projecting key ancient individuals from the region1,34,35,36,37,38. b, Shared genetic drift of present-day groups with the Leang Panninge individual, as calculated using f3 (Mbuti; Leang Panninge, X) mapped at the geographical position of the tested group. WGA, whole genome amplification. c, The amount of introgressed Denisovan sequence in fragments longer than 0.05 cM in present-day (Simons Genome Diversity Project) individuals and longer than 0.2 cM in ancient individuals (measured with admixfrog). Each bar represents the posterior mean estimate from a single genome and the whiskers indicate 2 s.d. (estimated from 200 samples from the posterior decoding).
The most distinctive archaeological assemblages associated with Holocene hunter-gatherers in Wallacea belong to the Toalean technocomplex (8–1.5 kya)3,4,27,28. Found only in a 10,000 km2 area of South Sulawesi3 (Fig. 1b), Toalean cultural assemblages are generally characterized by backed microliths and small stone projectiles (‘Maros points’)4 (Extended Data Fig. 1a–c). In 2015, excavations at Leang Panninge in the Mallawa district of Maros, South Sulawesi (Fig. 1b), uncovered the first relatively complete human burial from a secure Toalean context (Extended Data Figs. 1–5, Supplementary Information). The individual was interred in a flexed position29 in a rich aceramic Toalean stratum. Exposed at a depth of around 190 cm, the burial has an inferred age of 7.3–7.2 kyr cal BP obtained from 14C dating of a Canarium sp. seed (Extended Data Figs. 2, 3, Supplementary Table 1). Morphological characters indicate that this Toalean forager was a 17–18-year-old female with a broadly Australo-Melanesian affinity, although the morphology does not fall outside the range of recent Southeast Asian variation (Supplementary Information).
Genomic analysis
We extracted ancient DNA from bone powder obtained from the petrous portion of the temporal bone of the Leang Panninge individual. After library preparation, we used a DNA hybridization capture approach to enrich for approximately 3 million single-nucleotide polymorphisms (SNPs) across the human genome (1240K and archaic admixture capture panels30) as well as for the entire mitochondrial genome (mtDNA capture31). We retrieved 263,207 SNPs on the 1240K panel, 299,047 SNPs on the archaic admixture panel and the almost complete mtDNA sequence. Authenticity of the analysed ancient DNA was confirmed by short average fragment length, elevated damage patterns towards the molecule ends, and low autosomal and mtDNA contamination estimates (Supplementary Fig. 1). We confirmed that the individual was genetically of female sex. Analysis of the polymorphisms present in the reconstructed mtDNA sequence suggests a deeply divergent placement within mtDNA haplogroup M (Supplementary Table 17, Supplementary Fig. 2).
We initiated our genomic investigation by principal component analyses (PCAs), comparing the Leang Panninge genome with present-day individuals from East Asia, southeast Asia and Near Oceania (comprising Indigenous Australia, Papua New Guinea and Bougainville) genotyped on the Human Origins SNP panel18,32,33,34. The newly generated genome and relevant published genomes from ancient individuals from eastern Eurasia were then projected on the PCA1,34,35,36,37,38. Leang Panninge falls into PCA space not occupied by any present-day or ancient individuals, but is broadly located between Indigenous Australian peoples and the Onge (Fig. 2a, Extended Data Fig. 6). F3-statistics33 of the form f3 (Mbuti; Leang Panninge, X), where X is replaced with present-day Asian-Pacific groups, indicated that the new genome shares most genetic drift with Near Oceanian individuals (Fig. 2b). We confirmed these results with f4-statistics33, suggesting similar affinity of Leang Panninge and Papuan individuals to present-day Asian individuals, despite Near Oceanian groups forming a clade to the exclusion of Leang Panninge (Extended Data Fig. 7a, b). All present-day groups from the region, with the exception of the Mamanwa and the Lebbo26, carry only a minor contribution of Papuan-related ancestry (Supplementary Fig. 4).
Fig. 2: The Leang Panninge genome within the regional genetic context.

a, PCA calculated on present-day individuals from eastern Eurasia and Near Oceania, projecting key ancient individuals from the region1,34,35,36,37,38. b, Shared genetic drift of present-day groups with the Leang Panninge individual, as calculated using f3 (Mbuti; Leang Panninge, X) mapped at the geographical position of the tested group. WGA, whole genome amplification. c, The amount of introgressed Denisovan sequence in fragments longer than 0.05 cM in present-day (Simons Genome Diversity Project) individuals and longer than 0.2 cM in ancient individuals (measured with admixfrog). Each bar represents the posterior mean estimate from a single genome and the whiskers indicate 2 s.d. (estimated from 200 samples from the posterior decoding).