|
|
Post by Admin on Mar 14, 2020 4:56:08 GMT
Where did the Japanese come from? Based on their findings, the researchers came closer to answering the question of where the Japanese come from. The genetic information of the Jomon woman tells us that her ancestors descended from continental Eurasian people between 38,000 and 18,000 years ago. That means they had a longer period of hereditary isolation than previously assumed. 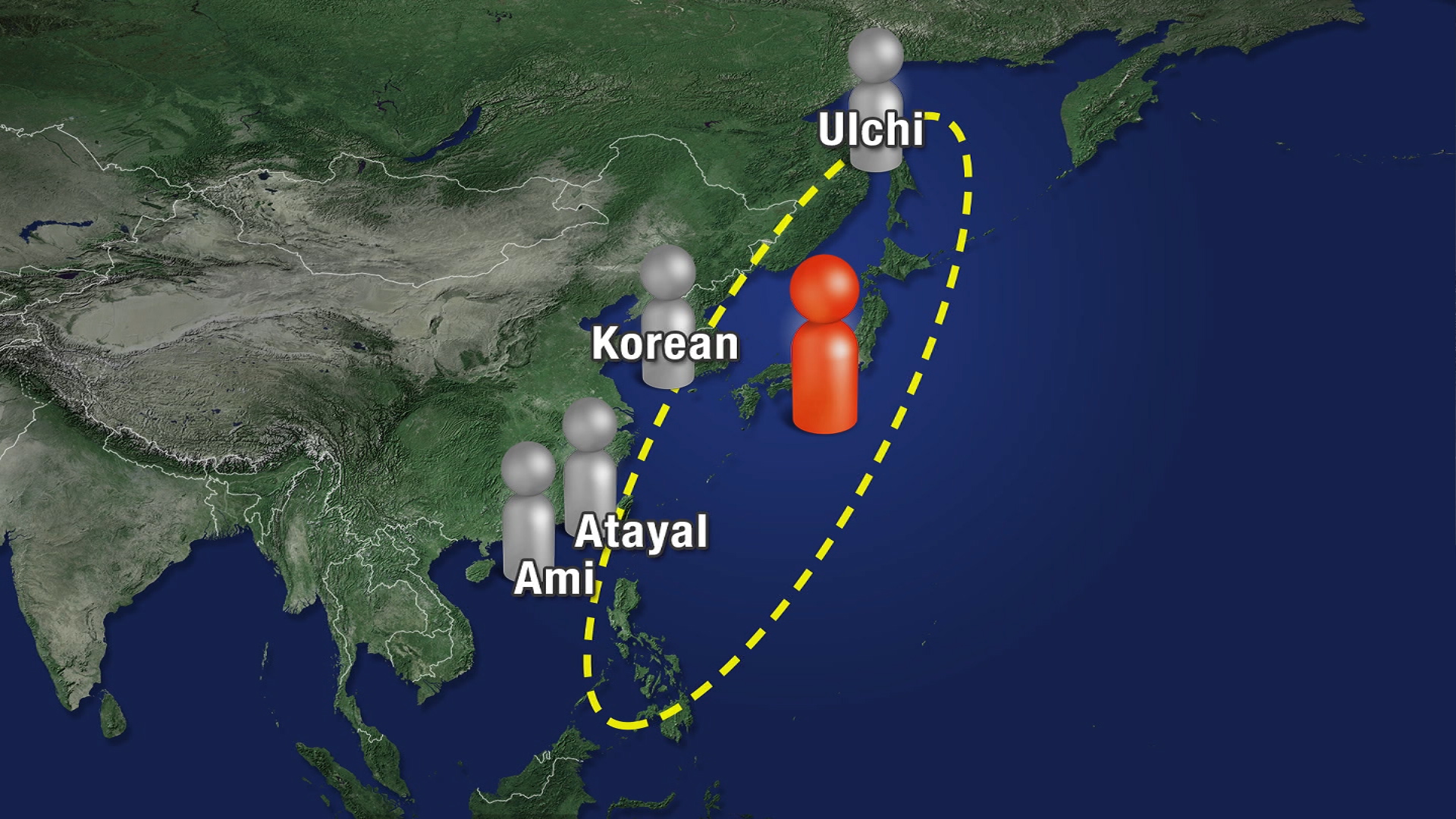 The research also showed that Jomon people shared genetic characteristics with people over a wide stretch of coastal East Asia, from north to south. The woman's genetic makeup has similarities with the Ulchi of the Russian Far East, as well as Koreans and the indigenous Taiwanese Atayal and Ami. These findings begin to add up to a profile of the ancestors of the modern Japanese. They suggest a higher genetic affinity with East Asian coastal peoples than with continental peoples like the Han Chinese. Kenichi Shinoda, Director of the Department of Anthropology at the National Museum of Nature and Science Kenichi Shinoda with the National Museum of Nature and Science has high hopes for the implications of the research. "I believe the study of the genes of prehistoric people is set to pick up and leap forward," he says. "The genetic information we have analyzed can hopefully contribute to unraveling the mystery of the origins of the Japanese, or to an understanding of the causes of the diseases to which the Japanese are prone." For the next phase of the project, researchers will more closely compare the DNA samples of the Jomon woman with modern Japanese over the next four years to find out what further secrets they reveal. We present 42 new Y-chromosomal sequences from diverse Indian tribal and non-tribal populations, including the Jarawa and Onge from the Andaman Islands, which are analysed within a calibrated Y-chromosomal phylogeny incorporating South Asian (in total 305 individuals) and worldwide (in total 1286 individuals) data from the 1000 Genomes Project. In contrast to the more ancient ancestry in the South than in the North that has been claimed, we detected very similar coalescence times within Northern and Southern non-tribal Indian populations. A closest neighbour analysis in the phylogeny showed that Indian populations have an affinity towards Southern European populations and that the time of divergence from these populations substantially predated the Indo-European migration into India, probably reflecting ancient shared ancestry rather than the Indo-European migration, which had little effect on Indian male lineages. Among the tribal populations, the Birhor (Austro-Asiatic-speaking) and Irula (Dravidian-speaking) are the nearest neighbours of South Asian non-tribal populations, with a common origin in the last few millennia. In contrast, the Riang (Tibeto-Burman-speaking) and Andamanese have their nearest neighbour lineages in East Asia. The Jarawa and Onge shared haplogroup D lineages with each other within the last ~7000 years, but had diverged from Japanese haplogroup D Y-chromosomes ~53000 years ago, most likely by a split from a shared ancestral population. This analysis suggests that Indian populations have complex ancestry which cannot be explained by a single expansion model. PMID: 28444560 DOI: 10.1007/s00439-017-1800-0 |
|
|
|
Post by Admin on Mar 14, 2020 21:27:40 GMT
Mitochondrial DNA analysis of the human skeleton of the initial Jomon phase excavated at the Yugura cave site, Nagano, Japan NOBORU ADACHI, JUNMEI SAWADA, MINORU YONEDA, KOICHI KOBAYASHI, SHIGERU ITOH Abstract Obtaining genetic information about early humans is indispensable to our understanding of the demographic history of mankind. In the present study, we performed a detailed mitochondrial DNA (mtDNA) analysis of a skeleton of the initial Jomon era unearthed from the Yugura cave site in Nagano, Japan, which was dated to 7920–7795 calBP by direct 14C dating. mtDNA of the Yugura skeleton was designated to haplogroup D4b2, which is widely observed in present-day East Asians, including the Japanese, but is absent in Hokkaido Jomon people. This finding indicates that the basal population of Japan was heterogeneous with respect to their mtDNA lineage. This is the first report on the genotype of the people from the initial phase of the Jomon period. 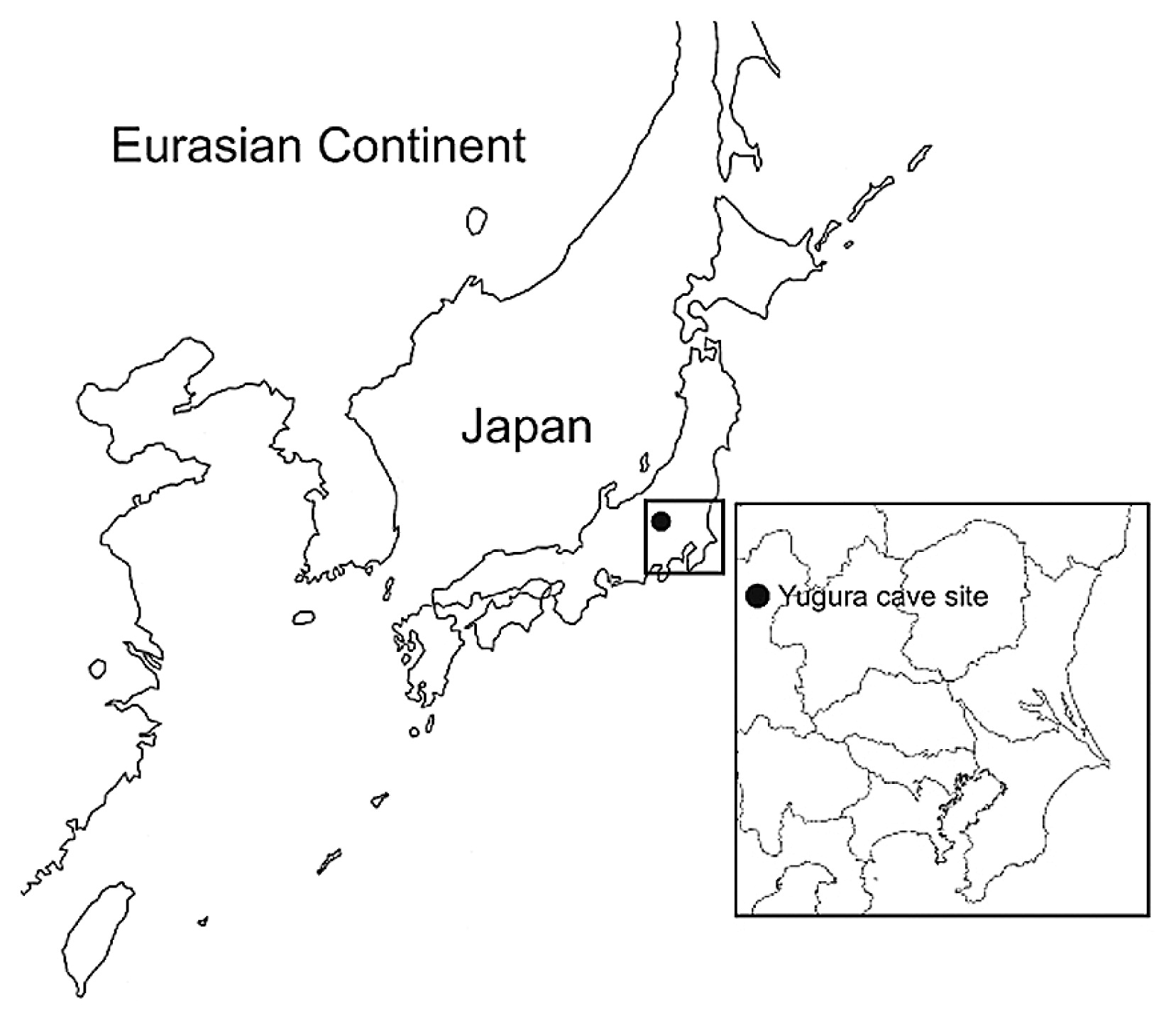 Figure 1 Map of the Yugura cave site, located in Nagano prefecture, central Japan. Introduction Mitochondrial DNA (mtDNA) studies of the modern Japanese have revealed that this population is rich in matrilineal diversity (e.g. Maruyama et al., 2003; Tanaka et al., 2004), and that there is considerable regional difference in haplogroup frequencies (e.g. Umetsu et al., 2005; Matsukusa et al., 2010). Although Jomon people are generally considered to be homogeneous from the morphological viewpoint (e.g. Hanihara, 1991), mtDNA analyses of the Jomon people suggest that maternal heterogeneity and its regional differences were observed as long ago as the Jomon era (Horai et al., 1989; Shinoda and Kanai, 1999; Shinoda, 2003; Adachi et al., 2011). Moreover, studies on lithic industries indicate that regional differences in their styles can be extended back to the Upper Paleolithic era (reviewed by Oda, 2003). This cultural variation may reflect the difference of the genetic background of the people. To clarify the origin of this heterogeneity, obtaining genetic information about the early (if at all possible, Paleolithic) humans is indispensable. However, currently, the oldest genetic data of humans in Japan are from early Jomon skeletons (about 6000 years before present) (Horai et al., 1989; Adachi et al., 2011). In the present study, we performed a detailed mtDNA and radiocarbon analyses of the Jomon skeleton unearthed at the Yugura cave site in Nagano, Japan. This skeleton belongs to the initial Jomon phase, and it is known to be one of the oldest Jomon skeletons in Japan. |
|
|
|
Post by Admin on Mar 14, 2020 23:59:52 GMT
Materials and Methods Skeletal material The Yugura cave site is located at an elevation of 1500 m, 1.5 km west of the Kenashi Pass in Takayama village, Nagano, Japan (Figure 1). Excavations at the site were carried out intermittently from 1971 to 1995. In August 1983, a well-preserved skeleton was unearthed from the excavation site at the location specified by grid C-3 of the site in Figure 2. Although the skeleton lacked grave goods, the whole body was found within layer IX (Figure 3), which was characterized by Oshigata-mon style pottery. Since layer VIII (just above layer IX) is estimated to belong to the initial Jomon era (Board of Education, Takayama village, 2001), this skeleton was presumed to originate from the middle of the initial Jomon era. 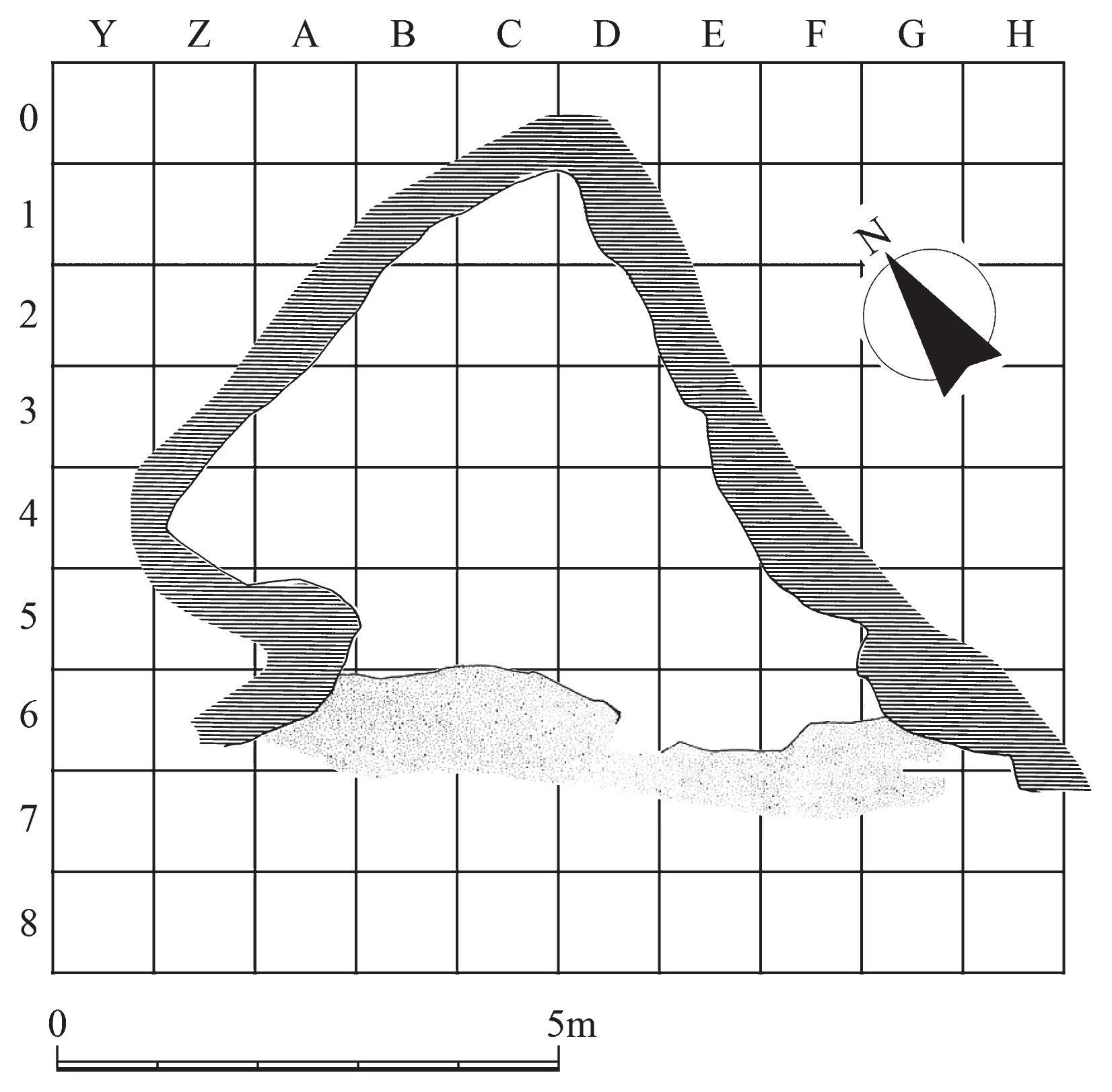 Figure 2 Floorplan of the Yugura cave site (from Board of Education, Takayama village (2001), modified). 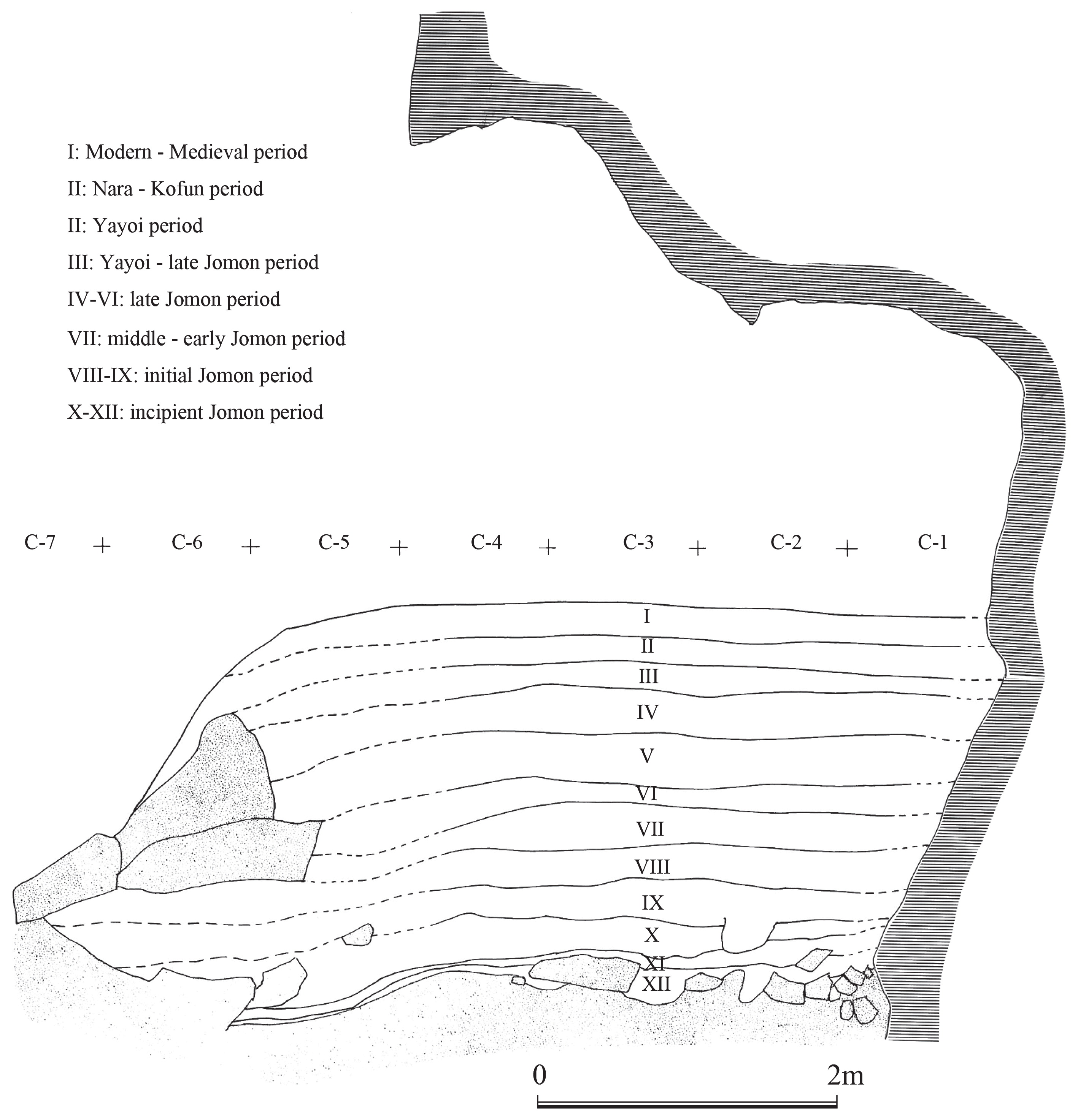 Figure 3 Layers of the Yugura cave site (from Board of Education, Takayama village (2001), modified). The skeleton was buried in the right lateral cramped position, and the direction of the head was facing northeast. Moreover, a stone was placed on the body (Figure 4, Figure 5). Based on the morphological evidence, the skeleton was estimated to be that of an adult female (Morimoto and Takahashi, 1986). 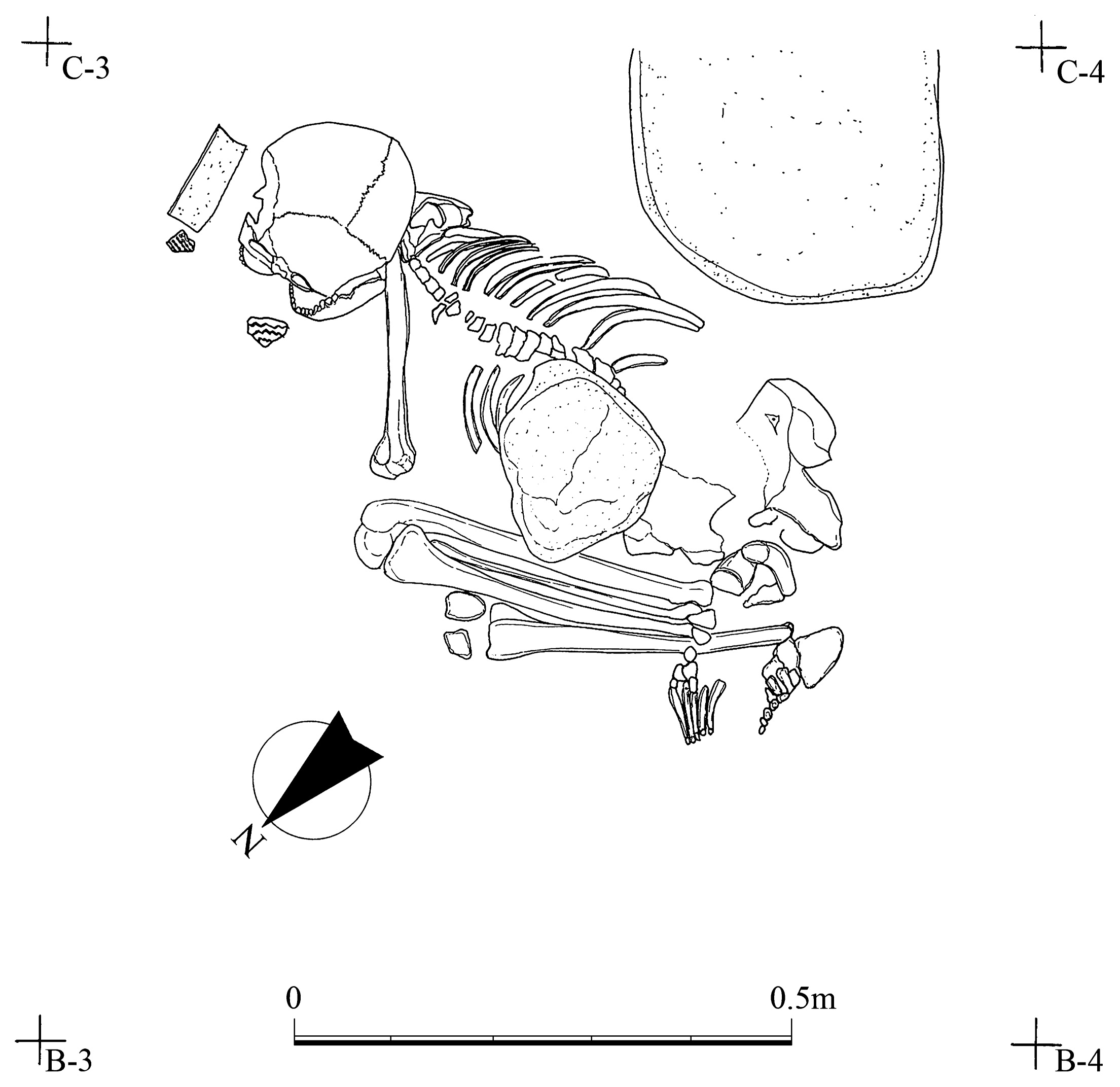 Figure 4 Plan of the skeleton excavated from the Yugura cave site (from Board of Education, Takayama village (2001), modified). |
|
|
|
Post by Admin on Mar 15, 2020 20:51:25 GMT
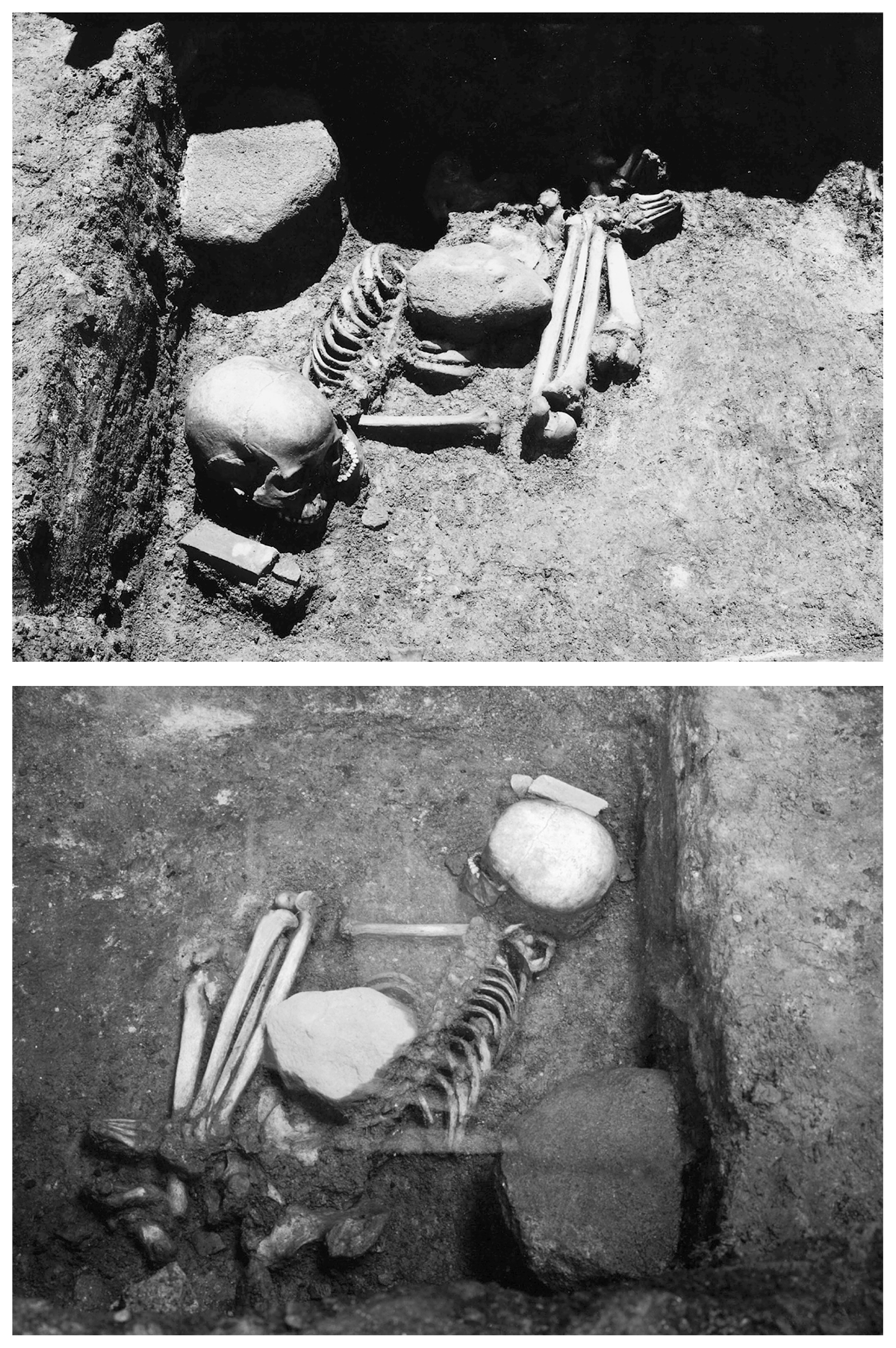 Figure 5 Picture of the skeleton excavated from the Yugura cave site. Radiocarbon dating Collagen was extracted by the gelatinization method (Longin, 1971; Yoneda et al., 2002), and its purity collagen estimated based on yield, carbon and nitrogen concentrations (Van Klinken, 1999). Radiocarbon dating was conducted on a graphite target produced from extracted collagen (Kitagawa et al., 1993) using the accelerator mass spectrometry (AMS) facility of Paleo Labo Co., Ltd. (Kobayashi et al., 2007). Extraction and purification of DNA Intensive precautionary measures to avoid modern DNA contamination were carried out as described previously (Adachi et al., 2011). DNA was extracted from the left lower third molar and from the left lower first molar independently, in order to validate the results. Tooth samples were dipped in a 13% bleach solution (Nacalai Tesque Inc., Kyoto, Japan) for 15 min, rinsed several times with DNase/RNase-free distilled water (Invitrogen, Carlsbad, CA), and allowed to air-dry. Furthermore, the outer surface of the samples was removed to a depth of 1 mm with a dental drill. Next, the samples were rinsed with DNase/RNase-free distilled water once again and allowed to air-dry under irradiation in a UV crosslinker for 90 min. The samples were then encased in Exafine silicone rubber (GC, Tokyo, Japan) following the protocol described by Gilbert et al. (2003). After approximately 10 min, or once the rubber was hard, the tip of the root of the tooth sample was removed with a horizontal cut and then powdered using a mill (Multi-beads shocker®; Yasui Kikai, Osaka, Japan). Thereafter, the dentin around the cavitas dentis and the dental pulp was powdered and removed through the root tip by using a dental drill. The powdered samples were decalcified with 0.5 M EDTA (pH 8.0) (Invitrogen) at 56°C overnight. The EDTA buffer was then replaced with a fresh solution, and the samples were decalcified for a further 36 h at 56°C. Decalcified samples were lysed in 1000 μl of Genomic Lyse buffer (Genetic ID, Fairfield, IA) with 50 μl of 20 mg/ml proteinase K (Invitrogen) overnight at 56°C. Finally, the lysate was extracted twice with 1200 μl of phenol/chloroform/isoamyl alcohol (25:24:1) and 1200 μl of chloroform. DNA was extracted from the lysate by using a FAST ID DNA Extraction Kit (Genetic ID) in accordance with the manufacturer’s instructions. Sequencing and single nucleotide polymorphisms typing of mtDNA Segments of mtDNA that covered parts of the tRNAPro gene, the hypervariable segments (HVS) 1 (nucleotide position (np) 15999–16141, 16121–16238, and 16209–16366, relative to the revised Cambridge reference sequence (rCRS: Andrews et al., 1999), HVS 2 (np 128–256), and a segment of the coding region that covered parts of the NADH dehydrogenase 3 and tRNAArg genes (np 10287–10425) were amplified and sequenced using primers that have been described previously (Adachi et al., 2009). In this regard, the –21M13 and M13RVN sequences were removed from these primers to maximize the robustness of the polymerase chain reaction (PCR). To exclude the possibility of modern DNA contamination, candidates for contamination were analyzed for the segments described above. Informed consent was obtained from the DNA donors. This study was approved by the ethics committee of the University of Yamanashi. Amplifications were carried out in a total reaction volume of 20 μl, containing a 2 μl aliquot of DNA extract, 0.2 μM of each primer, and reagents of the Multiplex PCR kit (QIAGEN, Hilden, Germany). The conditions for PCR included incubation at 95°C for 15 min, followed by 36 cycles at 94°C for 20 s, 54°C for 90 s, and 72°C for 15 s, and a final extension at 72°C for 10 min. Following PCR, the products were subjected to direct sequencing using the same primers and the BigDye® Terminators v. 1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on a 310 DNA Sequencer (Applied Biosystems) equipped with Ridom TraceEdit software (Ridom, Würzburg, Germany). To securely assign mtDNAs to relevant haplogroups, 36 haplogroup-diagnostic single nucleotide polymorphisms (SNPs) and a 9 bp repeat variation in the non-coding cytochrome oxidase II/tRNALys intergenic region (9 bp) were analyzed by the multiplex amplified product-length polymorphism (APLP) method. Protocols were identical to those described previously (Adachi et al., 2009), except for the PCR condition: incubation at 95°C for 15 min, followed by 40 cycles at 94°C for 30 s, 56°C for 90 s, and 72°C for 10 s, and a final extension at 72°C for 10 min. Thereafter, we assigned Yugura mtDNAs to the relevant haplogroup according to the HVS and coding region data, by using the classification tree built by Van Oven and Kayser (2009). |
|
|
|
Post by Admin on Mar 16, 2020 20:48:13 GMT
Results and Discussion Radiocarbon date The elemental analysis on extracted collagen showed a carbon concentration of 43.4% and a nitrogen concentration of 14.9%, indicating acceptable preservation status based on this C/N atomic ratio of 3.4 (acceptable range 2.93.6; DeNiro, 1985). The conventional radiocarbon age is 6999 ± 27 BP (PLD-21877), corresponding to 7920–7795 calBP (calculated by OxCAL 4.1 program and IntCal09 calibration dataset with one standard deviation; Bronk Ramsey, 2009; Reimer et al., 2009). This result shows that the skeleton analyzed here certainly originated from the initial Jomon period, generally assigned to 115007000 calBP (Kobayashi, 2008). mtDNA haplogroup of the Yugara skeleton The mtDNA data of the left lower third molar and the left lower first molar of the Yugura skeleton could be determined unambiguously and are shown in Figure 6 and Table 1. The mtDNA data derived from the two teeth were identical, and distinct from those of the candidates for modern DNA contamination. Therefore, the data was considered to be authentic and convincing. 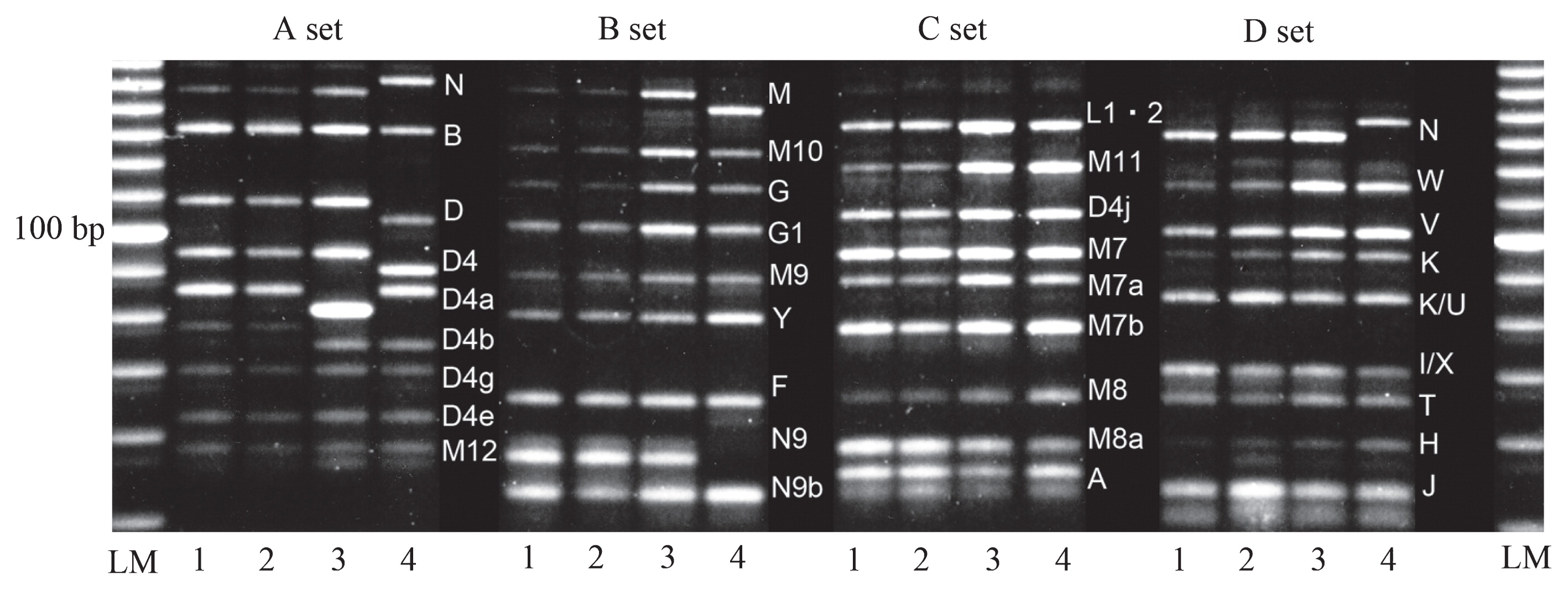 Figure 6 Haplogrouping of the samples by using four 9-plex APLP (A, B, C, and D set; Umetsu et al., 2005). Lane LM, 10 bp ladder (Invitrogen); lane 1, Yugura (left lower first molar); lane 2, Yugura (left lower third molar); lane 3, Modern sample (D4a); lane 4, Modern sample (N9a). 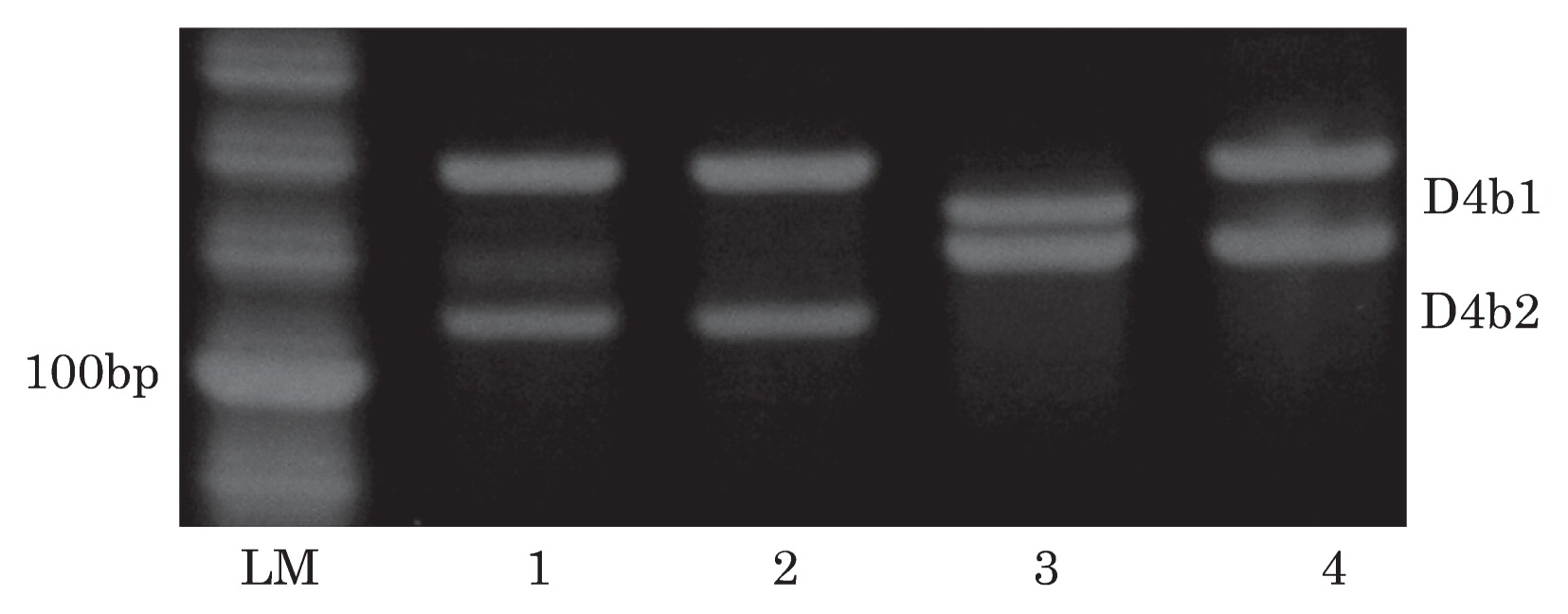 Figure 7 Subhaplogrouping of the samples obtained from the primer sets shown in Table 2. Lane LM, 10 bp ladder (Invitrogen); lane 1, Yugura (left lower third molar); lane 2, Yugura (left lower first molar); lane 3, Hatanai 35 (D4b1, Adachi et al., 2004); lane 4, Hatanai 30 (G1a, Adachi et al., 2004). Electrophoresis conditions are the same as described in Umetsu et al. (2005). Haplogroup D4b is widely observed among East Asian populations including Japanese populations (e.g. Yao et al., 2002; Umetsu et al., 2005; Lee et al., 2006; Nohira et al., 2010). Although the timing, origin, and route of migration of this haplogroup to the Japanese archipelago is still unknown, our study demonstrates that human beings with haplogroup D4b mtDNA were already present in the center of the Japanese archipelago during the initial phase of the Jomon era. Prehistoric anthropological significance and future research directions With regard to the early Jomon samples, the Hokkaido Jomon individuals contained haplogroups M7a (Kita-Kogane No. 7) and D4h2 (Kita-Kogane No. 9; Irie No. 12) (Adachi et al., 2011). Inferring from the HVS1 sequence motif (16223–16291–16362), another early Jomon mtDNA sample (Urawa 1: Horai et al., 1989) can be assigned to haplogroup E1a1a. The addition of haplogroup D4b detected in the Yugura skeleton indicates that at least four haplogroups were likely present by the early stage of the Jomon period. This finding implies that the early Jomon people were heterogeneous in terms of their mtDNA lineage. Interestingly, in the late Upper Paleolithic period, micro-blade cultures that were mainly distributed in the western part of Japan (e.g. the Yadegawa industry) spread to central Japan in conjunction with northern-style microblade culture (Oda, 2003; Ambiru, 2010; Inada, 2010; Tsutsumi, 2010, 2011). In addition, the lithic source area study indicated that there might have been a long-distance trade of raw materials in the Upper Paleolithic period (Suzuki, 1973a, b; Ono, 2007; Ambiru, 2010; Tsutsumi, 2011). These archaeological findings indicate that there might have been a wide range of human interactions accompanying the exchange of material cultures prior to the Jomon era. Therefore, it is possible that the heterogeneity of the early Jomon people might have arisen from that of the Upper Paleolithic people. Moreover, the presence of haplogroup E1a1a in Urawa 1 may hint at the fact that wide-scale human interaction was still observed in the Jomon era, since this haplogroup seems to date to the early Holocene (7200 years, 95% CI: 4600–9800 years) and to originate in Southeast Asia (Soares et al., 2009). However, to validate our hypothesis, a large number of human skeletons from the initial and early Jomon (preferably Paleolithic) period must be genetically analyzed. |
|









