|
|
Post by Admin on Jun 24, 2019 17:45:19 GMT
Archaeological studies have shown that the Japanese prehistory is divided into three periods: the Paleolithic period (older than 14,500 years before present [YBP]), the Jomon period (14,500-2,300 YBP), and the Yayoi period (2,300-1,700 YBP)1. One of the most important events in Japanese history is the transition from subsistence food production activities such as hunting and gathering to farming rice at the beginning of the Yayoi period. The advanced rice farming is thought to have been introduced to mainland Japan (we use the term “mainland Japan” to distinguish Honshu, Shikoku and Kyusyu from Hokkaido and Okinawa in this paper), inhabited by the Jomon people, by the Yayoi immigrants who came from the Asian continent. 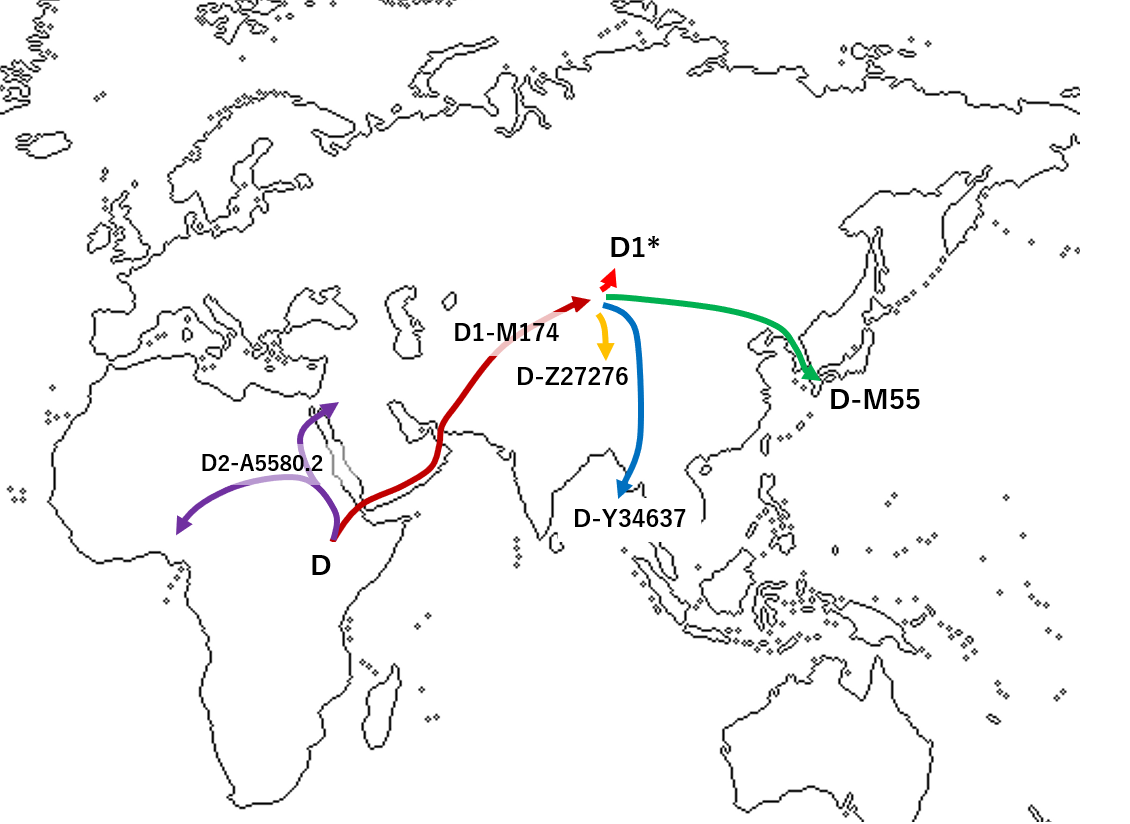 At present there are two minor populations and one major population in the Japanese archipelago: the Ainu who mainly live in Hokkaido (a minor population); the mainland Japanese (the major population); and the Ryukyuan who mainly live in Okinawa (a minor population). The prevailing hypothesis on the origins of the Japanese is “the dual structure model,2” where the present-day Japanese population is formed by an admixture between indigenous Jomon people and Yayoi immigrants who migrated from continental East Asia (Fig. 1). According to this model, modern mainland Japanese should have genomic components derived from both the Jomon people and those originating from the Yayoi immigrants. Genetic studies, using genome-wide single nucleotide polymorphism (SNP) data, basically supported the above model3,4,5,6. A recent study of ancient DNA extracted from the remains of Jomon individuals who had lived in mainland Japan (Fukushima Prefecture) 3,000 YBP suggested that (1) the Jomon people were strongly divergent from the current East Asians (Fig. 1), (2) the Ainu people are most closely related to the Jomon people among the three Japanese populations, and (3) ~12% of the genomic components of the modern mainland Japanese were derived from the Jomon people7. Genetically, the mainland Japanese are closely related to Koreans, followed by Han Chinese, and finally by other Continental East Asians3. These findings suggest that a large number of Yayoi immigrants came mainly through the Korean Peninsula to mainland Japan, and the mainland Japanese still retain genomic components from the Jomon people. Thus, there is a possibility that the historical change in the population size of the historic Jomon people, who lived in mainland Japan, could be estimated from genomic data from the modern mainland Japanese. 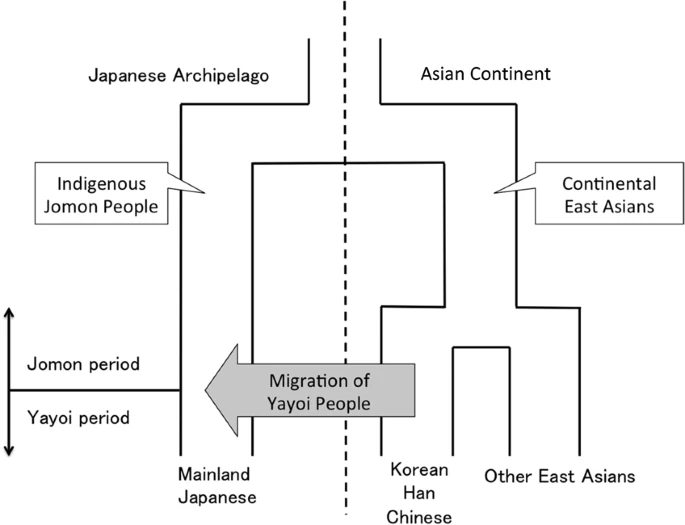 Figure 1 In the current work, we estimated the change in population size of the Jomon people in the Late to Final Jomon period by using whole Y-chromosome sequences of Japanese males living in mainland Japan. Specifically, we identified a phylogenetic clade (hereinafter called “clade 1”) specific to Japanese Y chromosomes and largely divergent from ones attributable to the continental East Asians. Y chromosomes in clade 1 appeared to be derived from the Jomon people. The population frequency of clade 1 Y chromosomes was estimated to be ~70% in Jomon males. The Bayesian skyline plot (BSP)8 for Y chromosomes in clade 1 allowed us to infer the historical change in population size of the majority of the Jomon people. Our results suggested that the Jomon people underwent a major decrease in population size at the end of the Jomon period, with a recovery occurring soon after the beginning of the Yayoi period. |
|
|
|
Post by Admin on Jun 25, 2019 4:51:35 GMT
Phylogenetic Tree of Y Chromosomes A total of 14 Mb of reliable sites had a call rate greater than 95% on the whole Y-chromosome sequences of 345 Japanese males (depth: 21×), and 28,254 Y-chromosomal SNPs were identified. Initially, a phylogenetic analysis was conducted for 345 Y chromosomes using the 28,254 SNPs. The Japanese Y chromosomes were divided into seven distinct clades in the neighbor-joining (NJ)9 tree (Fig. 2). To further investigate the phylogenetic relationship of East Asian Y chromosomes, a NJ tree of Y chromosomes from 345 Japanese, 47 Koreans10, and 244 East Asians11 was constructed using the 3,006 SNPs (Fig. S1). Although the branch lengths of the NJ tree in Fig. S1 were not correct because of the removal of population-specific variants, the topology was correct. We found a clade that contains only Japanese Y chromosomes in Fig. S1. Table 1 and Fig. 3 show the population frequency of each clade in East Asians. In this study, Japanese analyzed in 1000 Genomes Project are labeled “JPT” (“Japanese in Tokyo”) to distinguish our subjects from mainland Japanese. Incidentally, a total of 838 mitochondrial SNPs were identified from the same subjects (345 Japanese males). However, in contrast to the situation with the Y chromosomes, no clade specific to the Japanese population was found in the NJ tree of mtDNA sequences (Fig. S2). 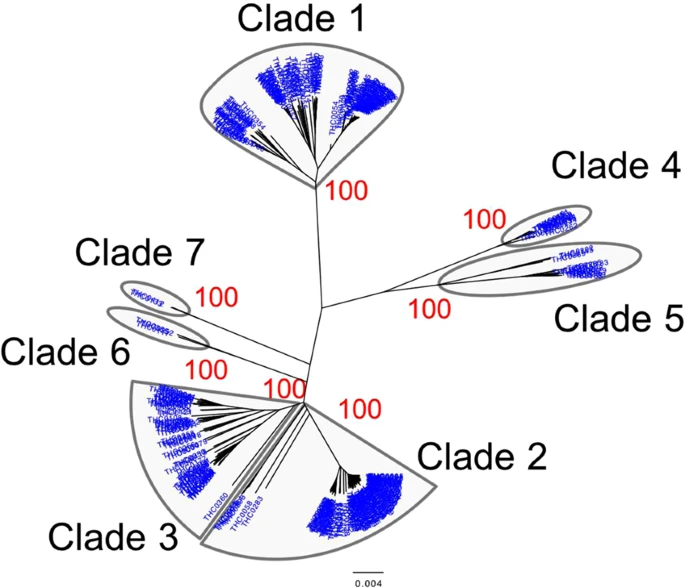 Figure 2 Y-Chromosome Clades in the Mainland Japanese The population frequency of Y chromosomes in clade 1 was 35.4% in the mainland Japanese and 35.7% in JPT (Table 1). The genotyping of rs2032626 (rs2032626-G allele is a marker for haplogroup D1b containing YAP+ variant) confirmed that clade 1 corresponded to haplogroup D1b (Fig. S3). Since the population frequencies of Y chromosomes in clades 2 and 3 were observed at relatively high frequencies in East Asians including Japanese (Table 1), a proportion of them seem to have been introduced to the mainland Japanese by Yayoi immigrants. The analysis of SNP markers revealed that Y chromosomes in clades 2 and 3 corresponded to haplogroups O1 and O2, respectively. Haplogroups O1 and O2 have been reported to be frequently observed in East Asians and Southeast Asians12. Clades 4 and 5 were assigned to haplogroup C, and clades 6 and 7 were assigned to haplogroup N. 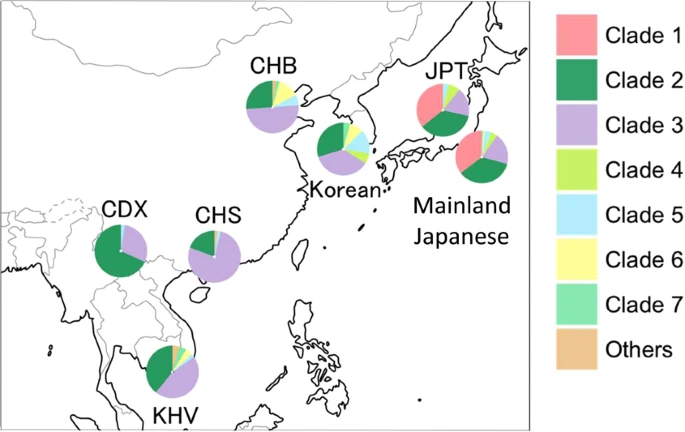 Figure 3 Frequencies of seven Y-chromosomal clades in East Asian populations. Y chromosomes in clade 1 were observed only in the Japanese among the East Asian populations. Frequencies of five East Asian populations were obtained from the 1000 Genomes Project phase 3 data. JPT: Japanese in Tokyo, Japan; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese; CDX: Chinese Dai in Xishuangbanna, China; and KHV: Kinh in Ho Chi Minh City, Vietnam. |
|
|
|
Post by Admin on Jun 26, 2019 18:02:22 GMT
Frequencies of Seven Y-Chromosome Clades in the Jomon People Y chromosomes in clade 1 were absent from continental East Asians except for Japanese (Table 1), and the frequency of haplogroup D1b (i.e. clade 1) was reported to be very high (81.3%) in the Ainu12, who have been shown, of the three Japanese populations, to be genetically and morphologically most closely related to the Jomon people7,13. These results strongly suggest that the mainland Japanese Y chromosomes in clade 1 originated from the Jomon people. The frequency of clade 1 in the mainland Japanese population (35.4% in the mainland Japanese and 35.7% in JPT) was much larger than the proportion (12%) of the genomic components of the Jomon people in the modern mainland Japanese, estimated previously by a study of ancient DNA from Jomon individuals7. Thus, the population frequency of clade 1 Y chromosomes must have been much higher in the Jomon people than in the present mainland Japanese. 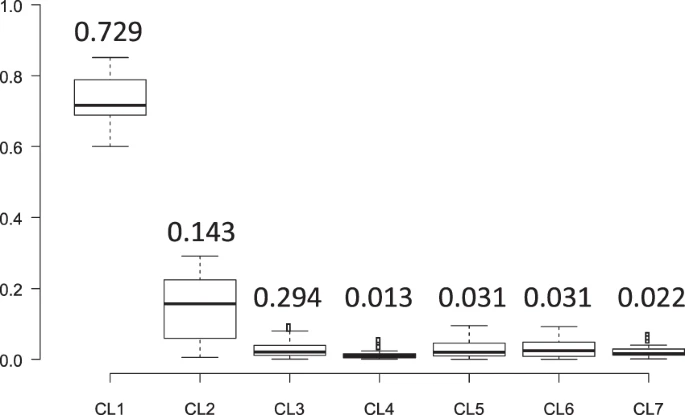 Figure 4 It was not possible to analytically estimate the population frequencies of seven clades in Jomon people. Instead, to confirm that the majority of Jomon people had clade 1 Y chromosomes, Monte-Carlo simulation, in which the population frequencies of the seven clades in the Jomon people are given by random numbers, was performed (see Material and Methods for details). The clade frequencies in an admixed population were calculated from those in the Jomon population (simulated) and those in Yayoi immigrants (assumed to be Koreans), and then compared with the clade frequencies observed in the current mainland Japanese (our subjects). The distribution of similarity index (SI) for 108 sets of the clade frequencies is shown in Fig. S4. In this analysis, SI decreased, as the difference between the simulated and observed clade frequencies in the mainland Japanese diminished. Only 20 sets (SI <3.24) were selected from 108 sets in order of increasing SI (Fig. 4), and the mean values of the population frequencies of Y chromosomes in clades 1, 2, and 3 were 73%, 14%, and 2%, respectively. This result implies that ~70% of the Jomon people in mainland Japan possessed clade 1 Y chromosomes, whereas the current mainland Japanese clade 3 Y chromosomes were mostly derived from Yayoi immigrants. 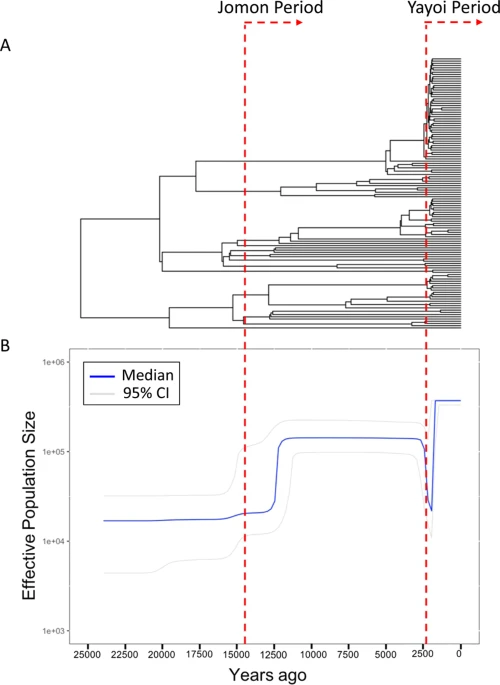 Figure 5 Bayesian Coalescent Inference of Historical Change in Population Size To deduce the historical change in the effective male population size of the Jomon people, Bayesian Skyline Plot (BSP) analyses8 were performed using BEAST 2.4.4 for 122 Y chromosomes of mainland Japanese belonging to clade 1 (Fig. 5). In the plot, the effective male population size started to increase gradually after the beginning of the Jomon period (12,500 YBP). Then, the population size decreased at 3,200 YBP (i.e. coalescent events frequently occurred 3,200 YBP as shown in Fig. 5A). Soon after the decrease, the effective population size of clade 1 rapidly increased to the present level. In addition, 68 Y chromosomes of the mainland Japanese in clade 3 were inspected to obtain information on the demography of a proportion of the Yayoi immigrants (Fig. S5). In contrast to the situation with clade 1, no population decrease was observed in the BSP of clade 3, implying that no severe bottleneck occurred when Yayoi immigrants came to the Japanese archipelago. This coincides with the previous observation that ~88% of the genomic components of the modern mainland Japanese were derived from the Yayoi immigrants7. |
|
|
|
Post by Admin on Jun 27, 2019 18:06:01 GMT
Coalescent Simulation In this study, only clade 1 Y chromosomes were used to infer the historical change in population size of Jomon people. Although ~70% of Jomon males are considered to have possessed clade 1 Y chromosomes, the use of such biased samples may prevent us from accurately estimating the time of an historical event. To confirm that the BSP result of the major clade (clade 1) reflects the time when the change in population size occurred, we performed coalescent simulations by fastsimcoal214, assuming that population size was changed three times (i.e., increase of 14,500 YBP, decrease of 3,000 YBP, and increase of 2,000 YBP). Then, the BSP was conducted for simulated chromosomes only in a major clade (frequency of more than 50%). The recent decrease and increase in the population size were successfully detected at the setting times (3,000 YBP and 2,000 YPB) in eight out of 10 simulation runs (Fig. 6, solid lines). As a consequence, the date at which the recent change in population size of Jomon population could be estimated by the BSP for Y chromosomes only in the major clade. However, considering the large difference in date of the population increase between the simulation results and the setting (14,500 YBP) in Fig. 6, the date of population increase around 12,500 YBP in Fig. 5 may not be reliable. 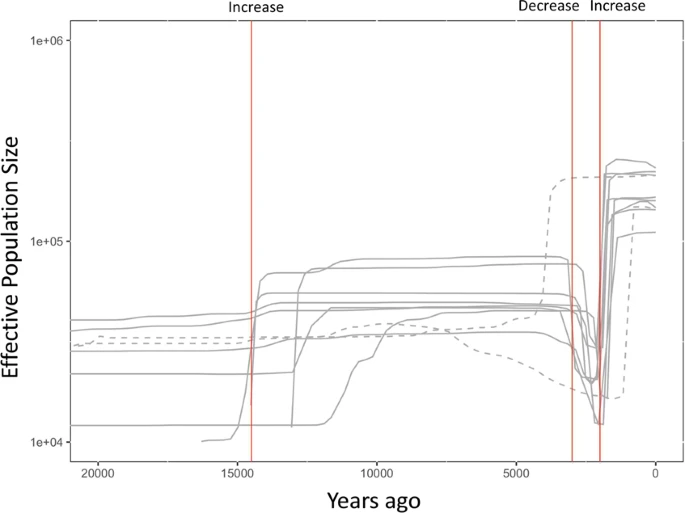 Figure 6 Discussion The present mainland Japanese have two ancestral populations: the indigenous Jomon and the Yayoi immigrants. In order to examine the demographic history during the Jomon period (14,500-2,300 YBP), we conducted a population genetic analysis using Y chromosomes of present-day mainland Japanese. In this study, a monophyletic group (named clade 1), consisting of only Japanese Y chromosomes, was identified in the phylogenetic analysis. Clade 1 was found to correspond to the Y– chromosome haplogroup D1b. This haplogroup, characterized by the insertion of an Alu element called YAP, is frequently observed in Japanese and Tibetans but is absent from other East Asians12,15. Although the lineages of haplogroup D1b in Japanese and Tibetans must have a common ancestor, the absence from Korean and Chinese populations suggests that the divergence of haplogroup D1b occurred before the ancestors of the Jomon people reached the Japanese archipelago. Y chromosomes in clade 1 are considered to have originated almost exclusively from the Jomon people who had inhabited the Japanese archipelago a long time before the Yayoi immigrants arrived. A simple computer simulation suggested that ~70% of the Jomon people carried the Y chromosome belonging to clade 1. In BSP for Y chromosomes in clade 1, we observed significant decreases in the effective population size of the Jomon people at the end of the Jomon period (Fig. 5). In the Final Jomon Period, there was a cooling trend in Japan with a sea level 1–3 m lower than the present level1. For the first time, we provide genetic evidence that the number of Jomon people decreased simultaneously with the cooling in the Final Jomon Period. Since the Jomon people were hunter–gatherers dependent on wild food, the number of the Jomon people might have decreased due to the shortage of food resources in the colder Final Jomon Period. A previous study16 had reported that a dietary change occurred along with climate change during the Late to Final Jomon period. It has been revealed that the density of archaeological sites declined in the Late to Final Jomon period17, suggesting that a sizeable population decline had occurred in Japan. Our results strongly support the archaeological finding. The effective population size of the Jomon people was estimated to have increased after the beginning of the Yayoi period (i.e., 2,300 YBP) (Fig. 5). The livelihood of the mainland Japanese shifted from nomadic hunter-gatherer to settled agriculturist in the Yayoi period. The rice farming introduced to mainland Japan by Yayoi immigrants, therefore, may have helped the Jomon people to procure food stably and recover their population size. Since it is known that effective male and female population sizes are different in general18 and the contributions of men and women are often unequal when admixture of two populations occurs19, the population history of Jomon females may be different from that of Jomon males. The ancient mtDNA analysis for Jomon people will add much to our understanding of historical changes in the effective female population size during the Jomon period. Scientific Reports volume 9, Article number: 8556 (2019) |
|
|
|
Post by Admin on Mar 14, 2020 1:30:54 GMT
 She had brown eyes, thin curly hair and dark skin that was prone to sun spots. Her earwax was wet and she had a high tolerance for alcohol, even though she never drank spirits. We know this and much more about a woman who lived and died 3,800 years ago. All of these details were deduced from her genetic information, meticulously pieced together by a Japanese research team in a breakthrough project of genome sequencing. The researchers mapped the entire genome of a person from prehistoric times to an unprecedented degree of precision and complexity. What they found tells us not just how she looked, but could eventually answer the question of where the Japanese came from tens of thousands of years ago, and how people lived on the archipelago in the millennia since. And given that modern Japanese inherited about 10 percent of the DNA present in the woman, she could contribute to an understanding of the diseases and medical conditions to which the Japanese are prone. 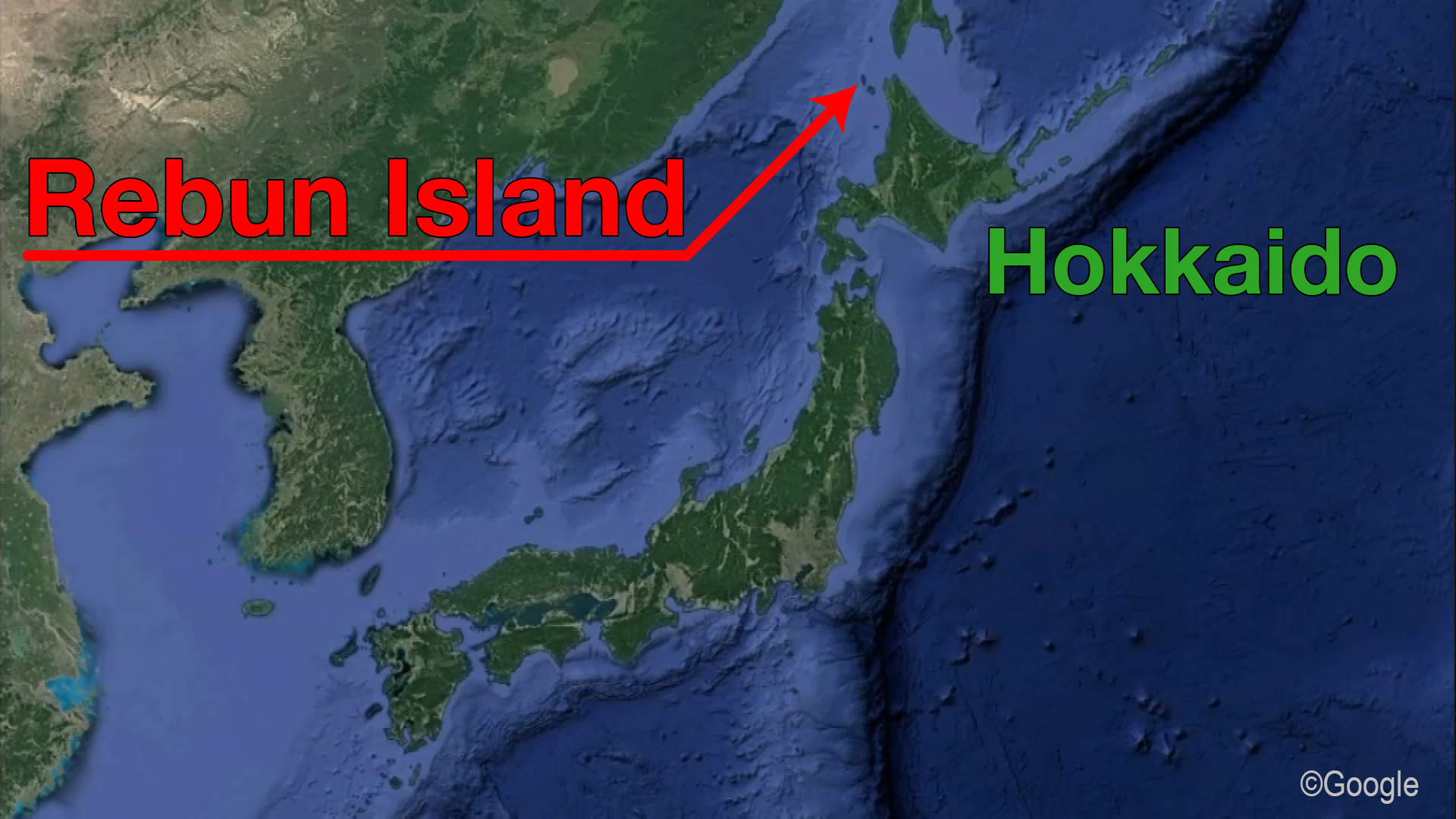 A woman from the past Researchers at the National Museum of Nature and Science and six other institutions launched an anthropological project to investigate the genetic origins of the Japanese. In May 2019, the findings were published in the journal Anthropological Science in a paper titled, "Late Jomon male and female genome sequences from the Funadomari site in Hokkaido, Japan." During Japan's Jomon period from about 16,000 years ago to 3,000 years ago, people lived as hunter-gatherers. As some of their DNA was passed down to modern Japanese, unraveling their genome is important to understand who the Japanese are and where they come from. This has only recently become possible with the rapid improvements in DNA sequencing technology. The woman lived on Rebun Island off the northern tip of Hokkaido, the northernmost prefecture of Japan. Her bones were excavated together with those of 27 of her fellow villagers. Researchers discovered that the mitochondria DNA contained in her remains, and those of another male, were in particularly good condition. The condition of such remains depends largely on the environment in which they were preserved. DNA deteriorates rapidly under warm conditions, so it was fortuitous that the woman came to rest in subarctic conditions. The researchers proceeded by taking a molar from her skull and drilling a hole to extract the nuclear DNA. The work was carried out in sterile conditions to prevent researchers from contaminating the sample with their own DNA. 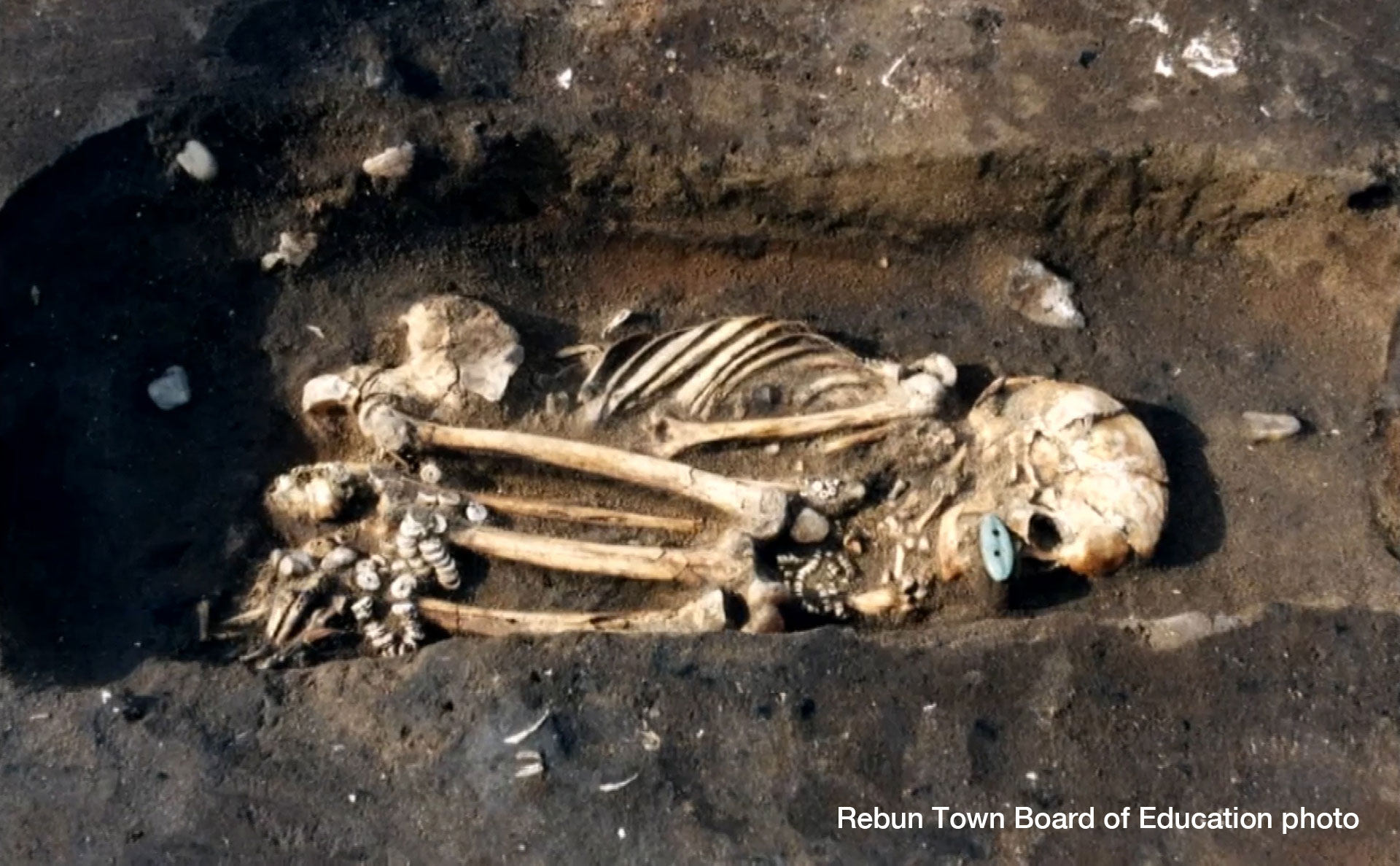 Previous work on sequencing the DNA of ancient human bones from other parts of Japan succeeded in mapping only some of the genome. This time, thanks to the perfect sample and the latest technological advances, the team teased out the genetic information of all 300 million pairs of the base sequence. Such complete and precise sequencing of ancient human DNA was an extraordinary feat. The researchers call it a "world first." The Jomon woman's skull is seen from different perspectives. In addition to determining many of the woman's physical characteristics, the analysis revealed that Jomon people had a genetic mutation involving amino acids that made it easier to metabolize fat. The characteristic is common among peoples in arctic regions, but doesn't exist among modern Japanese. It is considered proof that the Jomon people were hunter-gatherers. |
|












