|
|
Post by Admin on Apr 2, 2016 0:58:50 GMT
 Figure 1 More Recent Date of Denisovan than Neanderthal Admixture The Denisovans were named after an ancient human specimen found in the remote Denisova Cave in the Altai Mountains in Siberia. It was initially estimated that about 3% to 5% of the DNA of Melanesians and Aboriginal Australians has derived from Denisovans but this new study by Reich et al. (2016) revises their initial findings. Figure 2 shows that there are some hot spots in China and India. Some indigenous tribes with Oceanian ancestry may harbour a higher degree of Denisovan admixture than East and South Asians. 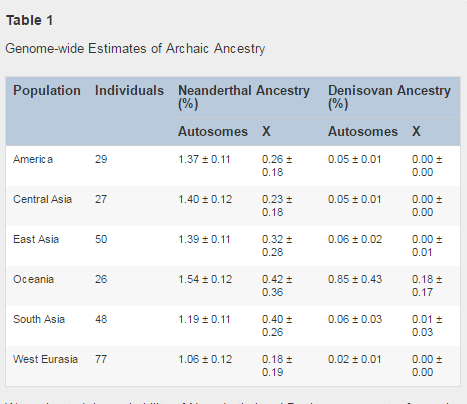 The percentage of Denisovan admixture in Oceania is 0.85 ± 0.43%, while the percentage is 0.06 ± 0.03% in South Asia, which are higher than other populations. The estimated Denisovan ancestry in East Asia, Central Asia, America are 0.06 ± 0.02%, 0.05 ± 0.01%, 0.05 ± 0.01%, respectively. Furthermore, the team led by David Reich previously argued that Denisovan ancestry was not detectable in West Eurasia (i.e. Europe) but the estimated figure is 0.02 ± 0.01% in West Eurasians and the fossil excavated in northern Spain called Sima de los Huesos was related to the Denisovans (Meyer et al. 2013).  Figure 2 Variation in Denisovan Ancestry Proportion For each individual, we inferred archaic ancestry segments across the autosomes (chromosomes 1–22) and chromosome X (our method did not allow us to test for archaic ancestry on chromosome Y because the archaic genomes are from females). Figure 2A plots the estimates of the proportion of confidently inferred Denisovan ancestry on a map, and Table 1 tabulates the results for six population pools (Table S2 tabulates the results for each population). Denisovan ancestry in Oceanians is greater than in other non-Africans [1] (Table 1). Both Neanderthal and Denisovan ancestry are greater in eastern non-Africans than in West Eurasians [6, 7, 8, 9, 10] (Supplemental Experimental Procedures, “Variation in the genome-wide proportions of archaic ancestry”; Table S3). We replicate previous findings of substantial Denisovan ancestry in New Guineans and Australians, as well as in populations that harbor admixtures of New Guinean ancestry [11]. However, we were surprised to detect a peak of Denisovan ancestry estimates in South Asians, both in the Himalayan region and in South and Central India (Figure 2A). The highest estimate is in Sherpas (0.10%), who have a Denisovan point estimate about one-tenth of that seen in Papuans (1.12%) (Table S3). Although this is notable in light of the likely Denisovan origin of the EPAS1 allele that confers high-altitude adaptation in Tibetans [12, 13], EPAS1 is not sufficient to explain the observation as Sherpas have the highest point estimate even without chromosome 2, on which EPAS1 resides. To determine whether the peak of Denisovan ancestry in South Asia is significant, we tested whether the Denisovan ancestry proportion in diverse mainland Eurasians can be explained by differential proportions of non-West Eurasian ancestry (as it is already known that there is more Denisovan ancestry in East Eurasians than in West Eurasians [6]). For each Eurasian population X, we computed an allele frequency correlation statistic that is proportional to eastern non-African ancestry (Figure 2B; Supplemental Experimental Procedures, “Modeling the variation in Denisovan ancestry across populations”). We regressed the proportion of confidently inferred Denisovan ancestry against this statistic. Although the proportion of Denisovan ancestry in these populations is correlated with non-West Eurasian ancestry (ρPearson = 0.832, block jackknife p = 3.6 × 10−10 for the correlation coefficient being non-zero), South Asian groups as a whole have significantly more Denisovan ancestry than expected (block jackknife Z score for residuals = 3.2, p = 0.0013 by a two-sided test for the null hypothesis that the Denisovan ancestry estimate in South Asians is predicted by their proportion of non-West Eurasian ancestry; Figure 2B; Supplemental Experimental Procedures, “Modeling the variation in Denisovan ancestry across populations”). The signal remains significant (Z = 3.1) when we remove from the analysis five populations that have ancestry very different from the majority of South Asians (Tibetan, Sherpa, Hazara, Kusunda, and Onge); however, the signals are non-significant for Central Asians (Z = 1.2) and Native Americans (Z = 0.1). Taken together, the evidence of Denisovan admixture in modern humans could in theory be explained by a single Denisovan introgression into modern humans, followed by dilution to different extents in Oceanians, South Asians, and East Asians by people with less Denisovan ancestry. If dilution does not explain these patterns, however, a minimum of three distinct Denisovan introgressions into the ancestors of modern humans must have occurred.  Figure 3 Fine-Scale Maps of Denisovan and Neanderthal Introgression One line of evidence for reduced fertility in male hybrids is that the proportion of archaic ancestry in modern humans is significantly reduced on chromosome X compared to the autosomes. This is suggestive of reduced male fertility as loci contributing to this phenotype are concentrated on chromosome X in hybrids of other species [22]. We confirm an extreme reduction of Neanderthal ancestry on chromosome X (16%–34% of the autosomes depending on the population) [14] and find a quantitatively similar reduction of Denisovan ancestry (21% of the autosomes in Oceanians) (Table 1). The second line of evidence in support of the hypothesis of reduced fertility in hybrids is that there is a reduction of archaic ancestry in genes that are disproportionately expressed in testes, a known characteristic of male hybrid fertility [22]. To test for this signal in our data, we analyzed a set of genes having a significantly higher expression level in testes than any of 15 others tissues in an RNA sequencing dataset [23]. We detect a statistically significant depletion of Denisovan (p = 1.21 × 10−7 in Oceanians) and Neanderthal (p = 2.1 × 10−3 in Oceanians) ancestry in these genes relative to the genes in the other tissues (Table S7; Supplemental Experimental Procedures, “Association of Denisovan ancestry with tissue-specific expression”). We considered the possibility that these observations could be explained by stronger linked selection at testes-expressed genes than at random places in the genome. However, when we correlate this pattern to B statistics (which are sensitive to linked selection [20]), we find that the gene sets that are disproportionally expressed in liver, heart, and skeletal muscle have even lower average B statistics than the genes most expressed in testes, and yet they do not show a depletion in archaic ancestry (Table S7). We also considered the possibility that the B statistic might not fully capture the degree of selective constraint at the genes disproportionately expressed in testes. However, when we use logistic regression to control for measures of selective constraint, we find that the significant reduction is observed not only when we control for B statistic at each gene (p = 4.4 × 10−7 for Denisovans; p = 2.8 × 10−3 for Neanderthals). It is also observed when we control for a direct estimate of the degree of selective constraint: the genetic diversity observed empirically at each gene in sub-Saharan Africans (p = 3.2 × 10−7 for Denisovans; p = 2.9 × 10−3 for Neanderthals; Supplemental Experimental Procedures, “Association of Denisovan ancestry with tissue-specific expression”). DOI: dx.doi.org/10.1016/j.cub.2016.03.037 |
|
|
|
Post by Admin on Jun 14, 2016 22:39:46 GMT
 The most mysterious interbreeding episode of all involves Denisovans and a ghost population of hominins that may have left Africa some million years ago. The mystery species could be an Asian offshoot of Homo erectus, which lived in Indonesia, perhaps as recently as 100,000 years ago, or possibly even relatives of Homo floresiensis, the 'hobbit' species discovered more than a decade ago on an Indonesian island.  An international team of researchers has just announced the discovery of 700,000-year-old remains that are related to a tiny species of human ancestors, lovingly referred to as 'hobbits'. Found on an island in Indonesia, the newly found hominins only grew to around 1 metre in height, and it's not year clear whether they represent a new species. 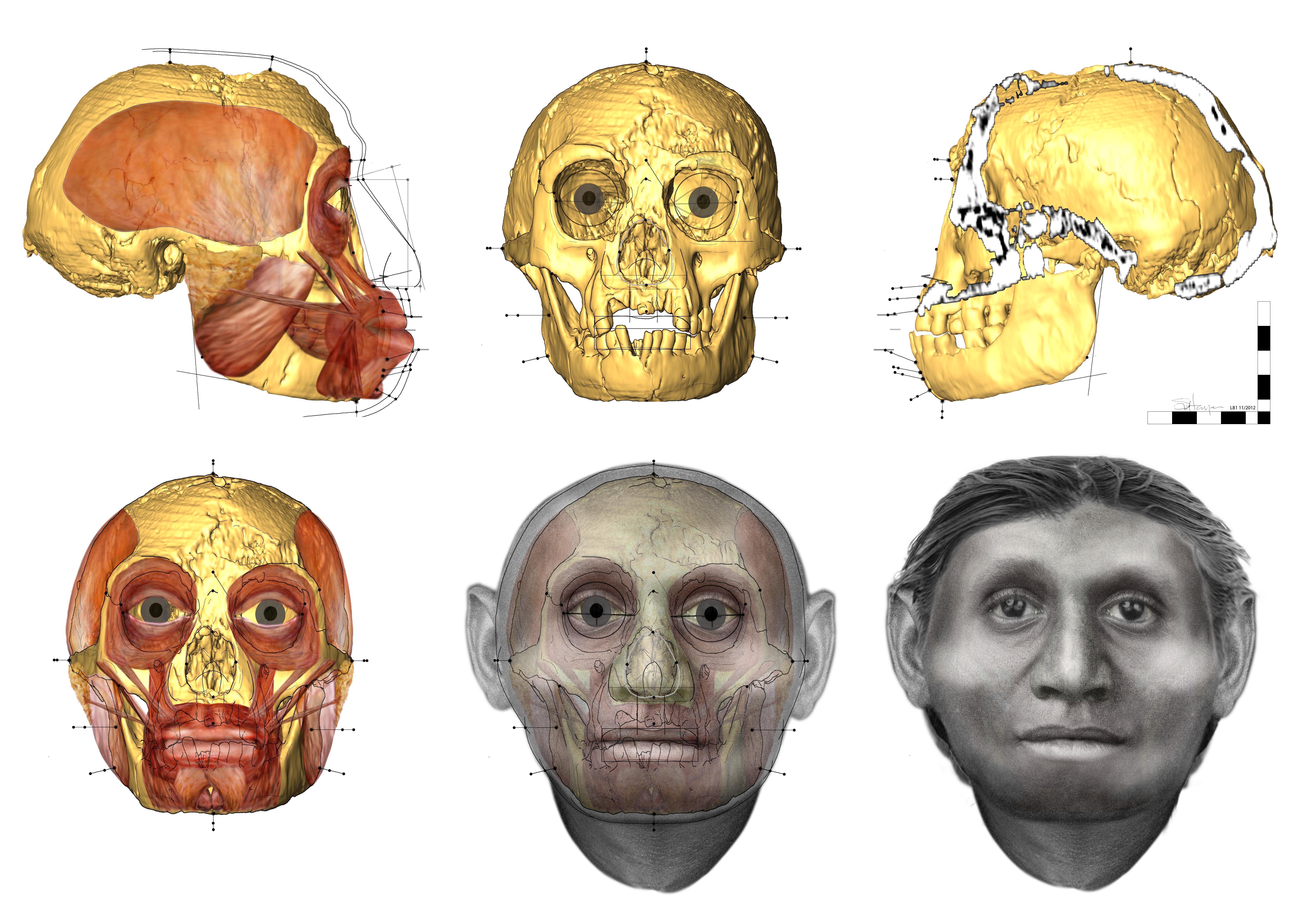 The team discovered a fragment of jaw and six teeth, from at least one adult and two children, buried beneath an ancient riverbed at a site known as Mata Menge, on the Indonesian island of Flores.  This is the same island where H. floresiensis was discovered back in 2003. Nicknamed the 'hobbit', the species is pretty incredible, because they might have lived alongside modern humans, right up until 12,000 years ago.  "All the fossils are indisputably hominin and they appear to be remarkably similar to those of Homo floresiensis," said one of the researchers, Yousuke Kaifu, from Tokyo's National Museum of Nature and Science, who compared the fossils against a modern and extinct hominins.  So where do these hobbits fit on our family tree? And how did they end up on a remote island? Publishing their results in Nature, the team proposes that they might have evolved from tall, upright species, Homo erectus, and somehow shrunk again.  Stone artefacts and fossils from Mata Menge. Recent excavations at the early Middle Pleistocene site of Mata Menge in the So’a Basin of central Flores, Indonesia, have yielded hominin fossils1 attributed to a population ancestral to Late Pleistocene Homo floresiensis2. Here we describe the age and context of the Mata Menge hominin specimens and associated archaeological findings. The fluvial sandstone layer from which the in situ fossils were excavated in 2014 was deposited in a small valley stream around 700 thousand years ago, as indicated by 40Ar/39Ar and fission track dates on stratigraphically bracketing volcanic ash and pyroclastic density current deposits, in combination with coupled uranium-series and electron spin resonance dating of fossil teeth. Palaeoenvironmental data indicate a relatively dry climate in the So’a Basin during the early Middle Pleistocene, while various lines of evidence suggest the hominins inhabited a savannah-like open grassland habitat with a wetland component. The hominin fossils occur alongside the remains of an insular fauna and a simple stone technology that is markedly similar to that associated with Late Pleistocene H. floresiensis.  To arrive at the 700,000-year figure for the bones’ age, the team used radiometric techniques to date volcanic layers above and below the soil layer where they were found, and also directly dated a partial hominin tooth. From abundant animal and plant remains they built a picture of the ancient environment: savannalike grasslands watered by meandering streams and populated by pygmy elephants, giant rats, freshwater crocodiles, and carnivorous Komodo dragons. They also examined 149 simple stone tools uncovered near the hominins, which are mostly similar to thousands uncovered elsewhere on Flores, says team member Mark Moore of the University of New England in Armidale, Australia. He notes that one kind of relatively complex tool characteristic of H. erectus appears in the Flores record only about 1 million years ago, and vanishes after that. But the other, simple tools, perhaps made by hobbitlike people, remain unchanged for hundreds of thousands of years. |
|
|
|
Post by Admin on Sept 10, 2016 21:17:55 GMT
 Figure 1: Ancestry of Indian populations. To shed light on the peopling of South Asia and the origins of the morphological adaptations found there, we analyzed whole-genome sequences from 10 Andamanese individuals and compared them with sequences for 60 individuals from mainland Indian populations with different ethnic histories and with publicly available data from other populations. 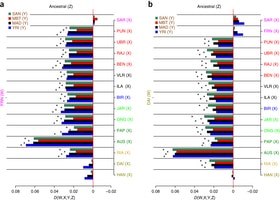 Figure 2: Fewer African-derived alleles in Indians, Andamanese, Papuans and Aboriginal Australians than in Europeans and East Asians. We show that all Asian and Pacific populations share a single origin and expansion out of Africa, contradicting an earlier proposal of two independent waves of migration1, 2, 3, 4. We also show that populations from South and Southeast Asia harbor a small proportion of ancestry from an unknown extinct hominin, and this ancestry is absent from Europeans and East Asians. The footprints of adaptive selection in the genomes of the Andamanese show that the characteristic distinctive phenotypes of this population (including very short stature) do not reflect an ancient African origin but instead result from strong natural selection on genes related to human body size.  Partha Majumder, director National Institute of Biomedical Genomics (NIBMG), said: “What we have been able to show is that there is strong possibility that there is at least one more ancestor and we have found the segments of this new ancestor, not found in either the Neanderthal or the Denisovan.  “Remains of this extinct hominid have not yet been recovered, but our results provide definitive evidence that Homo heidelbergensis had given rise to multiple lineages, not just the Neanderthal and the Denisovan. Nature Genetics (2016) doi:10.1038/ng.3621 |
|
|
|
Post by Admin on Oct 26, 2016 20:57:47 GMT
In 2003, scientists unearthed fossils in Liang Bua cave on the Indonesian island of Flores that belonged to an unknown hominin, a close relative of modern humans, that lived between 60,000 and 100,000 years ago. Scientists have suggested that this hominin was a unique branch of the human lineagenamed Homo floresiensis. Its diminutive 3-foot (1 meter) stature earned this hominin the nickname of the "hobbit," after the tiny folk in J.R.R. Tolkien's book of the same name.
Scientists have proposed that H. floresiensis evolved from a group of Homo erectus, an extinct human species that is the earliest undisputed ancestor of modern humans. Scientists also proposed that this population shrank in size either shortly before or after reaching Flores. Another possibility is that H. floresiensis evolved from even more primitive hominins with more ape-like skeletons and smaller brains, such as the extinct human species Homo habilis or even the prehuman Australopithecus species. Researchers have argued that if the hobbit did have such ancient origins, this would reveal that hominins left Africa much earlier than previously thought.
The fossils include an adult jaw fragment and six teeth from at least three individuals, including two tiny "milk teeth" from separate infants. The researchers found that these remains are at least 700,000 years old, dating to a time "when there were no modern humans on the planet," Yousuke Kaifu, a paleoanthropologist at Japan's National Museum of Nature and Science in Tokyo and co-lead author of one of the two studies, told Live Science. (Previous research has suggested that modern humans arose in Africa about 200,000 years ago.)
The researchers said the shape and age of the fossils suggest that these newfound hominins could be ancestors of H. floresiensis. "All the fossils are indisputably hominin, and they appear to be remarkably similar to those of Homo floresiensis," Kaifu said in a statement. Intriguingly, the recently unearthed Mata Menge fossils are significantly smaller than the previously discovered Liang Bua remains. For instance, the Mata Menge jaw fragment, which came from the lower jaw of an adult, is 20 percent smaller than the smallest H. floresiensis lower jaw from Liang Bua.
Radical changes in size are common when animals are trapped on islands. For instance, the extinct dog-size giant rats of East Timor are an example of island gigantism, while dwarf mammoths and dwarf dinosaurs are cases of island dwarfism. "To date, Flores is the only island in the world where we have fossil evidence for a human lineage evolving in isolation and adapting to an insular environment over a period of almost 1 million years," van den Bergh told Live Science. "That is the main reason why Homo floresiensis is so different from any other human lineage from Africa, Europe or mainland Asia."
|
|
|
|
Post by Admin on Nov 1, 2016 20:56:41 GMT
People from Melanesia, a region in the South Pacific encompassing Papua New Guinea and surrounding islands, may carry genetic evidence of a previously unknown extinct hominid species, Ryan Bohlender reported October 20 at the annual meeting of the American Society of Human Genetics. That species is probably not Neandertal or Denisovan, but a different, related hominid group, said Bohlender, a statistical geneticist at the University of Texas MD Anderson Cancer Center in Houston. “We’re missing a population or we’re misunderstanding something about the relationships,” he said.  This mysterious relative was probably from a third branch of the hominid family tree that produced Neandertals and Denisovans, an extinct distant cousin of Neandertals. While many Neandertal fossils have been found in Europe and Asia, Denisovans are known only from DNA from a finger bone and a couple of teeth found in a Siberian cave (SN: 12/12/15, p. 14). 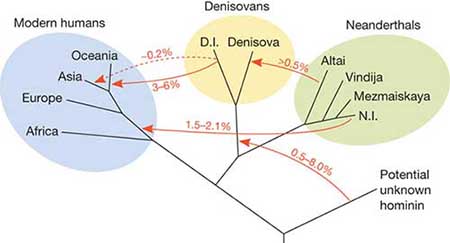 Bohlender isn’t the first to suggest that remnants of archaic human relatives may have been preserved in human DNA even though no fossil remains have been found. In 2012, another group of researchers suggested that some people in Africa carry DNA heirlooms from an extinct hominid species (SN: 9/8/12, p. 9). Less than a decade ago, scientists discovered that human ancestors mixed with Neandertals. People outside of Africa still carry a small amount of Neandertal DNA, some of which may cause health problems (SN: 3/5/16, p. 18). Bohlender and colleagues calculate that Europeans and Chinese people carry a similar amount of Neandertal ancestry: about 2.8 percent. Europeans have no hint of Denisovan ancestry, and people in China have a tiny amount — 0.1 percent, according to Bohlender’s calculations. But 2.74 percent of the DNA in people in Papua New Guinea comes from Neandertals, and another 3 to 6 percent stems from Denisovans, Bohlender calculated. Abstract: The sequencing of complete Neanderthal and Denisovan genomes has provided several insights into human history. One important insight stems from the observation that modern non-Africans and archaic populations share more derived alleles than they should if there was no admixture between them. We now know that the ancestors of modern non-Africans met, and introgressed with, Neanderthals and Denisovans. The estimate of the quantity of shared derived alleles, the mixture proportion, rests on an assumption of no archaic admixture in African populations, and so African populations have been used as the “non-admixed” outgroup in prior analyses. We find that the story is likely more complex, that the history within Africa involves admixture with population(s) related to Neanderthal and Denisova, and that the mixture proportion estimates for non-African populations have been biased, particularly in Melanesia. Here, we present results from a composite likelihood estimator of archaic admixture, which allows multiple sources of archaic admixture. We apply the method to archaic introgression, but it can be used to estimate ancient admixture among any four populations where the modeled assumptions are met. This joint estimate of Neanderthal and Denisovan admixture avoids the biases of previous estimators in populations with admixture from both Neanderthal and Denisova. To correct for dependence in our data, we use a moving blocks bootstrap to calculate confidence intervals. With assumptions about population size and more recent population separation dates taken from the literature, we estimate the archaic-modern separation date at ~440,000 ± 300 years ago for all modern human populations. We also estimate the archaic-modern mixture proportion in the 1000 genomes, and the modern genomes sequenced with the high coverage Neanderthal and Denisovan genomes. We report those estimates here, support several prior findings, and provide evidence for a lower level of Denisovan admixture (0.0191 [0.0184, 0.0197]), relative to Neanderthal (0.0256 [0.0247, 0.0265]), in Melanesia. On the basis of an excess of shared derived alleles between San, Neanderthal, and Denisova we suggest that a third archaic population related more closely to Neanderthal and Denisova than to modern humans introgressed into the San genomes studied here. |
|


















