|
|
Post by Admin on Jul 19, 2017 19:12:18 GMT
DNA retrieved from a child’s worn-down fossil tooth shows the ancient Asian roots of extinct Neandertal relatives called Denisovans, researchers say. A 10- to 12-year-old female Denisovan, represented by the tooth, lived at least 100,000 years ago, conclude evolutionary geneticist Viviane Slon of the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany, and colleagues. That makes the tooth at least 20,000 years older than the oldest of the three previously discovered Denisovan fossils — a finger fragment and two teeth — the scientists report July 7 in Science Advances. All four Denisovan specimens were unearthed in Siberia’s Denisova Cave.  Slon’s group extracted an almost complete set of mitochondrial DNA from the youngster’s tooth. Mitochondrial DNA typically passes from mothers to their children. Investigators also obtained a small amount of nuclear DNA, which is inherited from both parents. Comparisons of the ancient child’s DNA with that of the three other Denisovans, 10 Neandertals, five ancient humans, five present-day humans, a chimp and a roughly 400,000-year-old member of the Homo genus from Spain (SN Online: 3/14/16) enabled identification and approximate dating of the find. Previous dating of sediment layers in Denisova Cave helped to narrow down the fossil’s estimated age.  Despite Denisovans having inhabited the mountainous region around Denisova Cave for tens of thousands of years, the four fossil individuals attributed to the ancient population display relatively low levels of genetic diversity, the scientists say. Lack of diversity may be due to a small Denisovan population having inhabited one part of Asia for a long time. Or perhaps Denisovans lived elsewhere in Asia and evolved more genetic diversity than observed at the cave. The researchers say only discoveries of Denisovan fossils at additional sites can address that possibility. Abstract The presence of Neandertals in Europe and Western Eurasia before the arrival of anatomically modern humans is well supported by archaeological and paleontological data. In contrast, fossil evidence for Denisovans, a sister group of Neandertals recently identified on the basis of DNA sequences, is limited to three specimens, all of which originate from Denisova Cave in the Altai Mountains (Siberia, Russia). We report the retrieval of DNA from a deciduous lower second molar (Denisova 2), discovered in a deep stratigraphic layer in Denisova Cave, and show that this tooth comes from a female Denisovan individual. On the basis of the number of “missing substitutions” in the mitochondrial DNA determined from the specimen, we find that Denisova 2 is substantially older than two of the other Denisovans, reinforcing the view that Denisovans were likely to have been present in the vicinity of Denisova Cave over an extended time period. We show that the level of nuclear DNA sequence diversity found among Denisovans is within the lower range of that of present-day human populations. V. Slon et al. A fourth Denisovan individual. Science Advances. Published online July 7, 2017. |
|
|
|
Post by Admin on Jul 21, 2017 20:14:39 GMT
Mitochondrial DNA We used oligonucleotide probes matching a modern human mtDNA sequence to enrich for mtDNA fragments from the library that was not selected for uracil residues (4, 38). Initial inspection of the sequences suggested that the library contained a mixture of contaminating present-day human mtDNA and endogenous sequences that are more similar to a Denisovan mtDNA than to modern human or Neandertal mtDNAs (section S3 and table S4). We therefore aligned the sequences from the mitochondrial capture as well as DNA fragments sequenced without enrichment from all libraries to the Denisova 3 mtDNA genome (16) and identified 86,788 unique mtDNA fragments (table S2).  Fig. 1 Maximum likelihood tree relating the Denisova 2 mtDNA to other ancient and present-day mtDNAs. The Denisova 2 mtDNA (in red) clusters with the three previously determined Denisovan mtDNAs, to the exclusion of Neandertals and modern humans. Present-day human mtDNA sequences are noted in italics. The tree was rooted using a chimpanzee mtDNA sequence (not shown). Support for each branch is based on 500 bootstrap replications. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Accession codes for the comparative data and the geographical origins of ancient individuals are presented in table S5. To mitigate the influence of contamination by present-day human mtDNA (section S2), we filtered the 21,537 fragments (table S2) that carried C-to-T differences relative to the Denisova 3 mtDNA genome near the start or end position of sequence alignments (first three or last three positions for sequences from the standard library and first two or last two positions for sequences from the mini-U-selection protocol) (39). Using these fragments, we reconstructed the Denisova 2 mtDNA genome with an average mtDNA coverage of 51-fold (fig. S6). When a given position was required to be covered by at least three fragments and when at least two-thirds of fragments overlapping a position were required to carry an identical base (39), all but 14 positions in the mtDNA genome were resolved (section S3).  A maximum likelihood phylogenetic tree shows that the mtDNA of Denisova 2 clusters with the three previously determined Denisovan mtDNAs, to the exclusion of Neandertal and modern human mtDNAs (Fig. 1). It carries 29 nucleotide differences from Denisova 8, 70 nucleotide differences to Denisova 4, and 72 nucleotide differences to Denisova 3 (table S5). |
|
|
|
Post by Admin on Jul 23, 2017 19:11:00 GMT
Relative mtDNA dating The tree in Fig. 2 shows the relationships of the four currently known Denisovan mtDNAs using the Middle Pleistocene hominin mtDNA from Sima de los Huesos (39) as an outgroup. A total of 22.5, 33.5, 49.5, and 51.5 substitutions are inferred by parsimony (fractions reflect ambiguous character reconstructions) to have accumulated between the common ancestor and the mtDNAs of Denisova 2, 8, 4, and 3, respectively. In agreement with previous observations (17), Denisova 8 appears to be substantially older than Denisova 3 and 4. The mtDNA of Denisova 2 is more closely related to that of Denisova 8 than to the other two mtDNAs. Notably, 20 substitutions are inferred to have accumulated on the terminal edge leading to Denisova 8, whereas only 9 substitutions are estimated to have done so on the terminal edge leading to Denisova 2, indicating that Denisova 2 is older than Denisova 8.  Fig. 2 Phylogenetic tree relating the Denisova 2 mtDNA to other Denisovan mtDNA sequences. Using a mutation rate of 2.53 × 10−8 substitutions per site per year (95% highest posterior density, 1.76 × 10−8 to 3.23 × 10−8) (6), we estimate that the Denisova 2 individual is between 54.2 and 99.4 thousand years (ky) older than the Denisova 3 individual and between 20.6 and 37.7 ky older than the Denisova 8 individual, whereas the Denisova 3 and 4 individuals were roughly contemporaneous (between 3.7- and 6.9-ky difference). Although the absolute time estimates are dependent on whether the mutation rate of Denisovan mtDNA differs from that of modern human mtDNA, the difference in the number of substitutions between these individuals indicates that Denisova 2 is likely to be older than Denisova 8 and substantially older than Denisova 3 and 4. |
|
|
|
Post by Admin on Jul 25, 2017 19:48:44 GMT
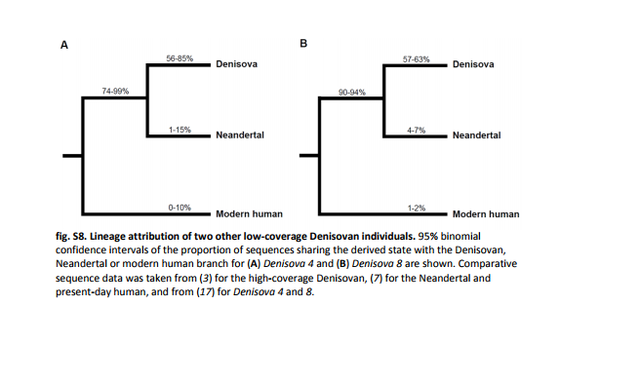 Attribution to a hominin group Genetic, archaeological, and anthropological evidence indicate that Denisovans, Neandertals, and anatomically modern humans were present in Denisova Cave (2, 3, 7, 16, 17, 28, 40, 41). We computed the proportion of DNA fragments from the Denisova 2 specimen that share derived alleles specific to each branch in a phylogenetic tree relating the high-coverage genomes of a Denisovan (3), a Neandertal (7), and a present-day human from Africa (7). A total of 69,315 fragments overlapped phylogenetically informative positions at which a randomly drawn allele from at least one of these high-coverage genomes is derived (12). Of fragments that overlap positions where both the Denisovan and the Neandertal genomes are derived, 84% (1888 of 2246) share the derived allele. Of fragments that overlap positions where only the Denisovan genome is derived, 49% (2051 of 4160) carry the Denisovan-like allele, whereas the corresponding values for the sharing of Neandertal- and modern human–specific alleles are 6% (252 of 4231) and 5% (307 of 5924), respectively (Fig. 3A). We thus conclude that the Denisova 2 specimen originated from a Denisovan individual.  Fig. 3 Attribution of Denisova 2 to a hominin group. (A) For each branch of a phylogenetic tree relating the high-coverage genomes of a Denisovan, a Neandertal, and a present-day human from Africa, the 95% binomial CIs of the proportion of DNA fragments from the Denisova 2 specimen that share a derived allele with that branch are given. (B) The fraction of substitutions inferred to have occurred after the split from the Denisova 2 genome along the branch from the human-chimpanzee (Ch) ancestral sequences to the high-coverage genomes of a Denisovan, a Neandertal, and 12 present-day humans (“X” in the schematic phylogenetic tree shown in the inset) is given. Error bars denote 95% CIs. The corresponding values for two previously sequenced Denisovan teeth, Denisova 4 and Denisova 8 (17), are 92 and 93% of sequences sharing derived alleles with the Neandertal-Denisovan branch, 72 and 60% with the Denisovan branch, 5% with the Neandertal branch, and 2% with the modern human branch, respectively (fig. S8). Thus, Denisova 2 shares fewer derived alleles with the high-coverage Denisova 3 genome than with the other two Denisovan genomes (χ2 = 7.6257, P = 0.003 and χ2 = 43.015, P = 2.717 × 10−11 for Denisova 4 and 8, respectively), showing that Denisova 2 is more distantly related to Denisova 3 than to Denisova 4 and Denisova 8. |
|
|
|
Post by Admin on Jul 27, 2017 19:11:11 GMT
 Denisovan DNA sequence diversity To gauge the average sequence divergence between the DNA fragments sequenced from Denisova 2 and the high-coverage Denisova 3 genome, we calculated how many of the substitutions on the lineage leading from the human-chimpanzee ancestor to Denisova 3 have occurred after the split from the Denisova 2 genome, that is, the fraction of derived sites that Denisova 2 does not share with Denisova 3 (1, 3, 7). This value is 5.9% [95% confidence interval (CI), 5.6 to 6.2%]. In comparison, it is 9.4% (95% CI, 9.0 to 9.9%) for derived alleles not shared with the Neandertal genome and 10.9 to 11.6% for 12 present-day human genomes (Fig. 3B and table S6). The corresponding values for Denisova 4 and Denisova 8 relative to the high-coverage Denisova 3 genome are 4.3% (95% CI, 2.3 to 6.7%) and 4.3% (95% CI, 4.0 to 4.7%), respectively. Excluding two Neandertal specimens yielding very little genetic data (0.1 Mb or less), the lower and upper boundaries of the 95% CIs for the fraction of substitutions occurring on the branch leading to the high-coverage Neandertal genome after the split from four low-coverage Neandertal genomes range between 2.6 and 4.2% (table S6). Comparable estimates calculated among 12 humans from various parts of the world range between 5.1 and 9.2% (table S7). Thus, the estimated sequence diversity among Denisovans, which all originate from a single cave, is comparable to that of Neandertals sampled in several locations and within the lower range of the diversity of present-day human populations worldwide. 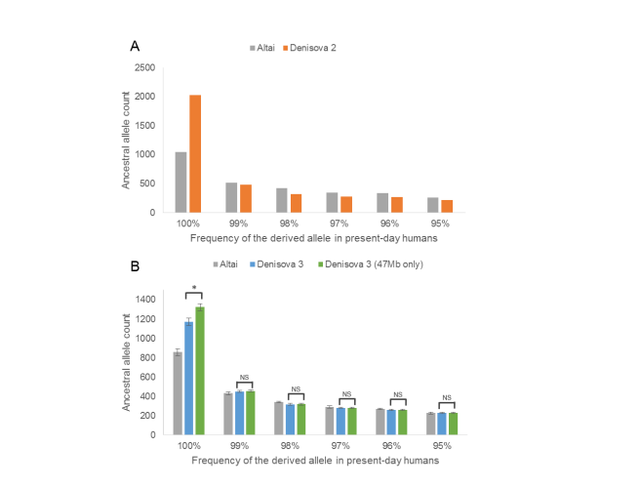 fig. S10. Ancestral allele counts in Denisovan and Neandertal sequences at sites that are fixed or nearly fixed for a derived allele in present-day humans. Denisova 2 was found in layer 22.1 of the Main Gallery of Denisova Cave, which has been dated to between 128 and 227 ka by radiothermoluminescence (42, 43). Denisova 3 was found in layer 11.2 of the East Gallery (16). Its age has been estimated to be between 48 and 60 ka on the basis of “missing substitutions” in its nuclear genome relative to present-day humans (7). This is consistent with the direct dating of associated finds, which are beyond the range of radiocarbon dating (2). Using nucleotide substitutions inferred to have occurred in the mtDNAs of Denisova 2 and Denisova 3 after their divergence from a common ancestor (Fig. 2), we estimate that the Denisova 2 individual lived approximately 50,000 to 100,000 years earlier than Denisova 3. Even given the uncertainty about the substitution rate in Denisovans, these results suggest that Denisova 2 lived at least 100 ka, making it one of the oldest hominin remains discovered in Central Asia to date. Beyond reinforcing the idea that both Neandertals and Denisovans lived in the cave or its vicinity (7, 17), our findings indicate that Denisovans were present over an extended period in the Altai region, where the two archaic groups may have met and mixed. The seemingly great difference in age between the Denisovan individuals is congruent with previous indications that Denisovans inhabited the Altai region over tens of thousands of years (17). Despite the wide age range of Denisovan individuals, their DNA sequence diversity is in the lower range of diversity among contemporaneous humans today. This low estimate of diversity among the Denisovans is consistent with the longtime small population size inferred from the high-coverage Denisova 3 genome (3). We note that age differences between the high-coverage archaic individuals and present-day modern humans can affect the comparability of our diversity estimates. However, this effect is expected to be minor, given that these branch length differences are far shorter than the common branch leading back to the human-chimpanzee ancestor. Moreover, as all four Denisovan individuals originate from a single location, it is possible that they represent an isolated population and that the genetic diversity of Denisovans across their entire geographical range was greater than that seen in these geographically restricted samples. Additional Denisovans from other locations are needed to more comprehensively gauge their genetic diversity across space and time. Science Advances 07 Jul 2017: Vol. 3, no. 7, e1700186 |
|









