|
|
Post by Admin on Aug 27, 2017 19:01:06 GMT
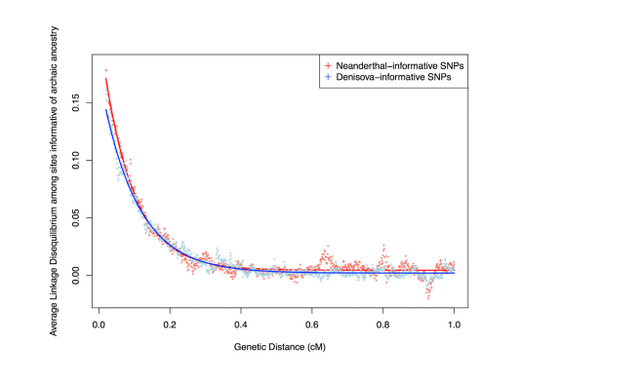 Figure 1. More Recent Date of Denisovan than Neanderthal Admixture Denisovan and Neanderthal Introgression Maps To study the impact of the Denisovan and Neanderthal admixture events simultaneously, we developed methods that allow us to distinguish these two sources of archaic ancestry. We applied these methods to the Simons Genome Diversity Project (SGDP) dataset: 257 high-quality genomes from 120 nonAfrican populations, including 20 Oceanian individuals from populations known to have high Denisovan admixture (unpublished data; Supplemental Experimental Procedures, ‘‘Data Processing’’). For each individual, we inferred archaic ancestry segments across the autosomes (chromosomes 1–22) and chromosome X (our method did not allow us to test for archaic ancestry on chromosome Y because the archaic genomes are from females). Figure 2A plots the estimates of the proportion of confidently inferred Denisovan ancestry on a map, and Table 1 tabulates the results for six population pools (Table S2 tabulates the results for each population). Denisovan ancestry in Oceanians is greater than in other non-Africans [1] (Table 1). Both Neanderthal and Denisovan ancestry are greater in eastern non-Africans than in West Eurasians [6–10] (Supplemental Experimental Procedures, ‘‘Variation in the genomewide proportions of archaic ancestry’’; Table S3). We replicate previous findings of substantial Denisovan ancestry in New Guineans and Australians, as well as in populations that harbor admixtures of New Guinean ancestry [11]. However, we were surprised to detect a peak of Denisovan ancestry estimates Figure 1. More Recent Date of Denisovan than Neanderthal Admixture Average linkage disequilibrium (Lewontin’s D) as a function of distance in Oceanians for SNPs informative of Neanderthal (red) and Denisovan (blue) ancestry. The Denisova decay is slower, implying a more recent date. See also Table S1. in South Asians, both in the Himalayan region and in South and Central India (Figure 2A). The highest estimate is in Sherpas (0.10%), who have a Denisovan point estimate about one-tenth of that seen in Papuans (1.12%) (Table S3). Although this is notable in light of the likely Denisovan origin of the EPAS1 allele that confers high-altitude adaptation in Tibetans [12, 13], EPAS1 is not sufficient to explain the observation as Sherpas have the highest point estimate even without chromosome 2, on which EPAS1 resides. 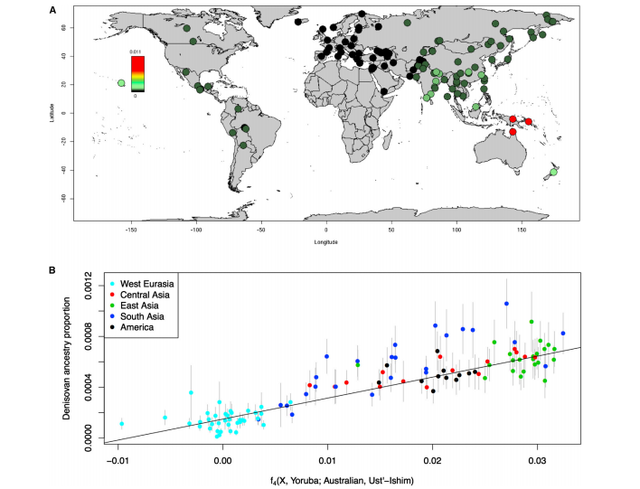 Figure 2. Variation in Denisovan Ancestry Proportion Tiling Path of Denisovan and Neanderthal Ancestry Inferred from Modern Genomes The union of detected Denisovan haplotypes spans 257 Mb in Oceanians (Supplemental Experimental Procedures, ‘‘Coverage of archaic haplotypes’’). The union of Neanderthal haplotypes spans 673 Mb over all non-Africans, which is smaller than the 1.1 Gb found in 1000 Genomes Project phase 1 data [14], most likely due to the fact that the total number of non-Africans genomes analyzed here is smaller. The positions of archaic ancestry are correlated across populations, with the strongest correlations at large spatial scales among the Neanderthal maps and weaker correlations between the Neanderthal and Denisovan maps (Figure 3B). Regions with Elevated Proportions of Archaic Ancestry We scanned all maps for windows with elevated proportions of archaic ancestry (average marginal probability R 0.30 over a 100 kb window based on a published threshold [4]; Supplemental Experimental Procedures, ‘‘Genomic regions with elevated archaic ancestry’’; Table S4). We identified 238 windows with elevated Neanderthal ancestry in a pool of all nonAfricans and 48 with elevated Denisovan ancestry in Oceanians. Regions with elevated archaic ancestry may represent loci where archaic alleles have experienced positive selection, but a formal test is challenging due to the fact that archaic alleles, on average, do not evolve neutrally [14–17]. We also tested for sets of genes that have among the 5% highest archaic ancestry (hypergeometric test implemented in FUNC [18]; we report p < 0.05 after multiple testing correction; Supplemental Experimental Procedures, section S4). Genes involved in keratin filament formation related to skin and hair are enriched for Neanderthal ancestry, generalizing the results of previous analyses that were limited to Europeans and East Asians [14, 15]. Genes involved in phospholipid transporter activity related to fat metabolism and in trace-amine receptor activity related to detecting subtle scents are significantly enriched for Denisovan ancestry (Table S5). |
|
|
|
Post by Admin on Aug 29, 2017 19:33:28 GMT
Deserts of Archaic Ancestry Some of the most striking features of the introgression maps are the archaic ancestry deserts: windows longer than 10 Mb at which the archaic ancestry proportion is <1/1000 (Figure 3A; Supplemental Experimental Procedures, ‘‘Analysis of genomic regions deficient in archaic ancestry’’). We identified 18 Neanderthal ancestry deserts in a pool of all non-African individuals and 63 Denisovan deserts in Oceanians. Four windows (1:99– 112 Mb, 3:78–90 Mb, 7:108–128 Mb, and 13:49–61 Mb) are both Neanderthal and Denisovan ancestry deserts. The desert on chromosome 7 contains the FOXP2 gene, which has been hypothesized to have a role in enabling modern human speech and language [19] and has been identified as a desert in previous maps in Europeans and East Asians. Our finding that this region is also a desert of Denisovan ancestry strengthens the evidence that the modern human version of this gene may be critical for modern human biology [14, 15]. Archaic Ancestry Is Reduced in the Genomic Regions Most Constrained by Selection We tested the relationship between archaic ancestry and regions of strong linked selection as measured by a B statistic [20]. Neanderthal ancestry decreases in proximity to functional elements in all populations (rSpearman = 0.25–0.29; Figure 3C; Supplemental Experimental Procedures, ‘‘Correlation of archaic ancestry with B-statistics’’; Table S6), as does Denisovan ancestry in Oceanians (rSpearman = 0.26, Table S6), most likely re- flecting greater selection against Neanderthal ancestry in low B statistic regions [14–17]. Power to detect archaic ancestry is elevated close to regions of linked selection due to a reduction in the rates of incomplete lineage sorting caused by the lower effective population size in these regions [14], so these results are not artifacts of reduced power. Thus, similar processes appear to have worked to remove Neanderthal and Denisovan ancestry near genes. 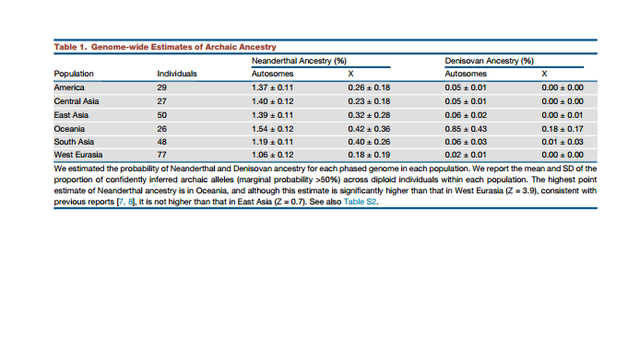 Archaic-Modern Admixture Was Most Likely Associated with Reduced Male Fertility Our study provides new evidence in support of the hypothesis that reduced male fertility may be a common feature of admixture between human populations diverged by at least a half million years, a hypothesis that was previously suggested based on genetic patterns associated with the hybridization between Neanderthals and modern humans [14, 21]. We show that qualitatively similar signals are associated with Denisovan admixture. One line of evidence for reduced fertility in male hybrids is that the proportion of archaic ancestry in modern humans is signifi- cantly reduced on chromosome X compared to the autosomes. This is suggestive of reduced male fertility as loci contributing to this phenotype are concentrated on chromosome X in hybrids of other species [22]. We confirm an extreme reduction of Neanderthal ancestry on chromosome X (16%–34% of the autosomes depending on the population) [14] and find a quantitatively similar reduction of Denisovan ancestry (21% of the autosomes in Oceanians) (Table 1)  It has been suggested that the empirically observed reduction in Neanderthal ancestry in Europeans and East Asians near functionally important regions could be explained by a greater load of weakly deleterious alleles in Neanderthals due to the smaller population size of Neanderthals since separation, followed by purging of deleterious Neanderthal alleles in the mixed population [16, 17]. Since we have shown that similar patterns are associated with the Denisovan introgression event, it seems plausible that similar evolutionary forces operated to remove Denisovan ancestry segments. However, the model of a greater load of deleterious mutations in archaic humans cannot explain the observed reduction of both Neanderthal and Denisovan ancestry near genes that are disproportionately expressed in testes, suggesting that male hybrid sterility may have been associated with both introgressions. An important direction for future research is to understand the relative importance of purging of slightly deleterious alleles, as well as reduced fertility in hybrid males, in changing the content of genomes in the aftermath of the interbreeding that occurred between modern and archaic humans. Sankararaman et al., The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans, Current Biology (2016) |
|
|
|
Post by Admin on Jan 18, 2018 19:12:47 GMT
 Following the initial description and analysis of a genome sequence from an archaic human fossil from Denisova Cave in southern Siberia (Reich et al. 2010), Denisovan admixture was subsequently found to be limited to populations from eastern Indonesia, the Philippines, and Near and Remote Oceania (Reich et al. 2010, 2011; Meyer et al. 2012). This finding was quite surprising, given that the Denisova Cave site is located some 7,000 km away from the populations that currently exhibit Denisovan ancestry, and was interpreted as suggesting that Denisovan admixture occurred somewhere in the vicinity of island Southeast Asia (Reich et al. 2011). However, further studies have indicated that Denisovan ancestry may be more widespread than initially thought (Skoglund and Jakobsson 2011). In particular, a recent study inferred low levels of Denisovan ancestry of about 0.2% in three genome sequences, from a Dai and a Han Chinese from East Asia and a Karitiana from South America (Prüfer et al. 2014). Moreover, it appears that a genetic adaptation to high altitude in the Tibetan Plateau occurred through introgression from a Denisovan-related population into ancestral Tibetans (Huerta-Sánchez et al. 2014). Overall, these results suggest that Denisovan ancestry is not limited to populations from island Southeast Asia and Oceania, as originally thought. However, only a few populations have been systematically evaluated for signals of Denisovan ancestry; it is not clear whether a very low level of Denisovan ancestry is geographically widespread, or rather limited to only a few populations outside of island Southeast Asia and Oceania. An additional question of interest is whether the Denisovan ancestry in these other populations reflects the same admixture event that contributed Denisovan ancestry to island Southeast Asian and Oceanian populations, or a different admixture event. To address these and other questions related to Denisovan ancestry in human populations, we here present a systematic investigation of Denisovan introgression in Eastern Eurasia (defined here to include South Asia, East Asia, Southeast Asia, Central Asia, Siberia, and Oceania) and in Native American populations, hereafter abbreviated as EE/NA. Genome-wide data were collected from worldwide populations and analyzed along with high coverage genome sequences from a Denisovan (Meyer et al. 2012) and a Neanderthal (Prüfer et al. 2014). The analyses we report provide more details concerning the admixture history of modern humans with Denisovans. 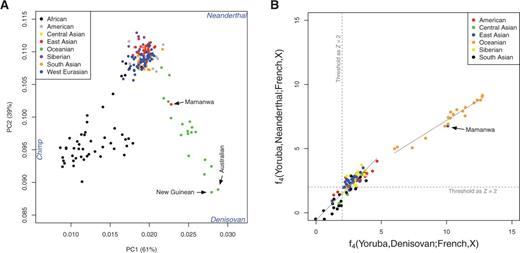 FIG. 1. The relationships among modern human populations relative to archaic humans. (A) PCA of 221 populations projected onto the top two eigenvectors defined by Neanderthal, Denisovan, and chimpanzee. The mean values of eigenvectors 1 and 2 are plotted for each population. (B) Formal admixture tests suggest a substantial number of EE/NA and Oceanian populations inherited significantly more archaic ancestry than West Eurasian populations. We used f4f4 statistics of the form f4(Yoruba,Archaic;French,X)f4(Yoruba,Archaic;French,X) to test admixture between archaic humans and modern human populations. A Z-score larger than 2 was set as the threshold for determining whether admixture is significantly greater than zero. We assembled a data set which, after quality filtering, consisted of 2,493 individuals from 221 populations (supplementary tables S1 and Supplementary Data, Supplementary Material online), all genotyped on the Affymetrix Human Origins Array (Patterson et al. 2012). After merging the human data with the chimpanzee, Denisovan, and Neanderthal genome sequences, there were nearly 600,000 single nucleotide polymorphisms (SNPs) for analysis. To investigate the relationship of the diverse present-day human populations relative to archaic hominins, we carried out Principal Component Analysis (PCA) (Price et al. 2006) on the chimpanzee, Neanderthal, and Denisovan data, and projected the modern human samples onto the plane defined by the top two eigenvectors. The human samples all appear at the center of the plot (supplementary fig. S1A, Supplementary Material online); magnification of the central portion of the plot shows that humans separate into three clusters relative to archaic hominins and chimpanzees: African, Oceanian, and other Non-African (supplementary fig. S1B, Supplementary Material online). To more clearly visualize the patterns, we plotted the mean of eigenvectors 1 and 2 for each of the 221 modern human populations (fig. 1A). The first eigenvector separates the Africans from non-Africans and shows that the non-Africans are clearly closer to archaic hominins than are the Africans. The second eigenvector suggests closer genetic affinity between Oceanians and Denisovan than between other populations and Denisovan. There is a clear cline of Denisovan-related ancestry in Oceanians with Australians and New Guineans having the most Denisovan ancestry (supplementary fig. S2, Supplementary Material online). The Mamanwa, from the Philippines, are also involved in this cline, which is consistent with previous findings that the Mamanwa are related to Australians and New Guineans, and Denisovan admixture occurred in a common ancestral population of the Mamanwa, Australians, and New Guineans (Reich et al. 2011). |
|
|
|
Post by Admin on Jan 19, 2018 19:01:50 GMT
Additional Archaic Ancestry in EE/NA Populations PCA is a descriptive analysis that is useful for indicating potential admixture events, but cannot be used to prove that admixture occurred. We therefore applied formal tests to document potential admixture between archaic hominins and modern humans. As EE/NA populations have on average inherited more archaic ancestry than West Eurasian populations (Wall et al. 2013), we computed f4f4 statistics (Reich et al. 2009; Patterson et al. 2012) of the form f4(Yoruba,Archaic;French,X)f4(Yoruba,Archaic;French,X) , in which X is an EE/NA population. A significantly positive statistic (Z-score >2>2 ) is evidence that EE/NA possesses more Archaic (either Neanderthal or Denisovan) alleles than does the French population. Significantly positive statistics (Z-score >6>6 ) are obtained for all Oceanian populations, which are much higher than those for other EE/NA populations, indicating more archaic ancestry in Oceanians (fig. 1B and supplementary table S3, Supplementary Material online). Moreover, there are more Denisovan than Neanderthal alleles shared with the Oceanian and Mamanwa populations (fig. 1B), although Neanderthal ancestry is also elevated, probably because signals of Denisovan and Neanderthal ancestry are difficult to distinguish in this analysis. In addition to the Oceanian populations, many additional EE/NA populations exhibit significant Z-scores ( >2>2 ), indicating that they have more archaic alleles than the French population has. However, unlike the Oceanian populations, the inferred amounts of Denisovan and Neanderthal alleles are approximately the same in these EE/NA populations (fig. 1B). It is thus not clear from this analysis whether the additional archaic ancestry in these EE/NA populations reflects Neanderthal ancestry, Denisovan ancestry, or both. In order to increase the power of these tests, we combined the data from the East Asian, Siberian, and native American populations, and obtained significantly higher signals of archaic ancestry (Z-score = 3.12 for Neanderthal and 3.64 for Denisovan, compared with the average single population Z-scores of 2.79 ± 0.07 for Neanderthal and 3.17 ± 0.08 for Denisovan). Although gene flow between the modern human reference populations could inflate the f4 statistics, the magnitude of the inflation is expected to be small relative to any archaic ancestry (see supplementary text S1, Supplementary Material online). To ensure that our results are not significantly influenced by the choice of African (Yoruba) and European (French) reference populations used in the f4f4 analyses, we repeated the analysis with different reference populations and found no significant differences in the results (supplementary fig. S3, Supplementary Material online). Moreover, values of f4f4 statistics are perfectly correlated with each other using different pairs of reference populations (supplementary fig. S4, Supplementary Material online). 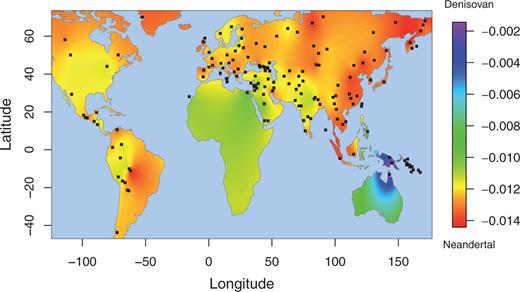 FIG. 2. Archaic introgression in modern humans is prevalent and varies across different geographic regions. The sharing of Neanderthal and Denisovan alleles with each non-African population was measured by f4f4 statistics of the form f4(Yoruba,X;Neanderthal,Denisovan)f4(Yoruba,X;Neanderthal,Denisovan) . An excess of allele sharing with Denisovan yields positive values, whereas an excess with Neanderthal yields negative values. The heat plot values indicated on the map are valid only for regions covered by our samples. We further computed the statistic f4(Yoruba,X;Neanderthal,Denisovan)f4(Yoruba,X;Neanderthal,Denisovan) which compares the genetic affinity of present-day non-Africans with different archaic hominins (fig. 2). Positive values of this f4 statistic indicate excess sharing of Denisovan alleles (relative to Neanderthals); negative values indicate excess sharing of Neanderthal alleles, and values near zero indicate equivalent amounts of alleles shared with Denisovans and Neanderthals. We obtain larger values in Oceanians than in other non-Africans, which is consistent with the observations based on the PCA and on formal tests of admixture. The largest values are observed in New Guineans, Australians, and some populations from Remote Oceania (supplementary table S4, Supplementary Material online), consistent with previous results (Reich et al. 2011). Negative values are observed in most non-African populations, indicating sharing of more Neanderthal than Denisovan alleles in these populations. Moreover, larger values are observed in East Asians than in West Eurasians, suggesting that most East Asians share more alleles with Neanderthal than West Eurasians do. Native Americans are generally more similar to West Eurasians in the patterns of allele sharing, suggesting that either Native Americans have the same amount of allele sharing with Neanderthal as West Eurasians do, or Native Americans shared more Neanderthal alleles than West Eurasians and additionally share a small amount of Denisovan alleles. This procedure was repeated using different African populations in the f4f4 statistics and similar results were obtained |
|
|
|
Post by Admin on Jan 20, 2018 19:12:48 GMT
 Denisovan Ancestry in Oceanians Denisovan ancestry in Oceanians has been documented previously (Reich et al. 2010, 2011; Meyer et al. 2012). To verify and extend these previous results, we analyzed a larger set of Oceanian populations that were genotyped on a different platform, and also utilized the high-coverage archaic genomes (Meyer et al. 2012; Prüfer et al. 2014). Following previous methods (Moorjani et al. 2011; Patterson et al. 2012), we used a ratio of f4f4 statistics to estimate the admixture proportion of Denisovans ( PD(X)PD(X) ) in Oceanians (see Materials and Methods). As Oceanians retain both Denisovan and Neanderthal ancestry, we used Han Chinese to control for the Neanderthal ancestry in Oceanians, under the assumption that Han and Oceanians share a similar number of Neanderthal alleles. To evaluate the validity of this assumption, we examined the f4(Yoruba,Han;Neanderthal,X)f4(Yoruba,Han;Neanderthal,X) and f4(Yoruba,Han;Denisovan,X)f4(Yoruba,Han;Denisovan,X) statistics for each Oceanian population X. If Han and the X possess similar amounts of Neanderthal alleles, then any changes in the two statistics will be driven by the varying amount of their Denisovan alleles. The two statistics will thus have a linear relationship with an intercept close to (0,0). However, if the amount of Neanderthal alleles differs in the two populations, then the linear model will not cross at the point of origin. Empirically, these two sets of f4f4 statistics are indeed linearly correlated ( R2=0.99; R2=0.99; supplementary fig. S6, Supplementary Material online) and the intercept for the linear model fitting the data is near (0,0), indicating that Han and Oceanians are similarly close to Neanderthal (see Materials and Methods).  FIG. 3. Denisovan ancestry in Oceanians. (A) Estimated Denisovan ancestry in Oceanian populations. (B) Denisovan ancestry in Oceanians is highly correlated with their New Guinean ancestry. The highest PD(X)PD(X) value is observed in Australians and New Guineans (0.034 ± 0.002 and 0.034 ± 0.005, respectively) (fig. 3A), which is consistent with previous results (Reich et al. 2011) and suggests Denisovan introgression into the common ancestor of Australians and New Guineans. We also observed high amounts (>3%) of Denisovan ancestry in Bougainville, as observed previously (Reich et al. 2011), and in  Cruz, a population from Remote Oceania which was not analyzed previously. This latter result is in keeping with previous observations of extraordinarily high frequencies and diversity of mtDNA and Y-chromosome haplogroups of New Guinean origin in  Cruz (Delfin et al. 2012; Duggan et al. 2014), which suggest high amounts of New Guinean ancestry (and thereby Denisovan ancestry) in  Cruz. All other Oceanian populations have Denisovan ancestry ranging from 0.9% to 3%. It was shown previously that Denisovan ancestry in Oceanian groups (other than Australia and New Guinea), as well as in eastern Indonesia, was likely to be an indirect consequence of admixture with New Guineans, as the Denisovan ancestry in these other Oceanian and eastern Indonesian groups is proportional to their New Guinea ancestry (Reich et al. 2011). We observe a similar correlation for the 17 Oceanian populations (excluding Australia and New Guinea) in this study (fig. 3B). However, given that Australia and New Guinea share common ancestry, it is possible that the Denisovan ancestry in these Oceanian populations was contributed by admixture from Australia or from the ancestral Australia–New Guinea population, rather than directly from New Guinea; these possibilities were not examined in previous studies. We evaluated these alternative possibilities by the statistic f4(Yoruba,X;NewGuinea,Australia)f4(Yoruba,X;NewGuinea,Australia) , and found that Oceanians share significantly more alleles with New Guineans than with Australians (supplementary table S5, Supplementary Material online). These results suggest that Denisovan ancestry in Oceanians is likely to derive from recent admixture with New Guineans, rather than admixture with Australians or the common ancestor of Australians and New Guineans. |
|










 Cruz, a population from Remote Oceania which was not analyzed previously. This latter result is in keeping with previous observations of extraordinarily high frequencies and diversity of mtDNA and Y-chromosome haplogroups of New Guinean origin in
Cruz, a population from Remote Oceania which was not analyzed previously. This latter result is in keeping with previous observations of extraordinarily high frequencies and diversity of mtDNA and Y-chromosome haplogroups of New Guinean origin in