Post by Admin on Dec 4, 2020 3:49:49 GMT
Section 5: Phenotypic analysis
Pigmentation
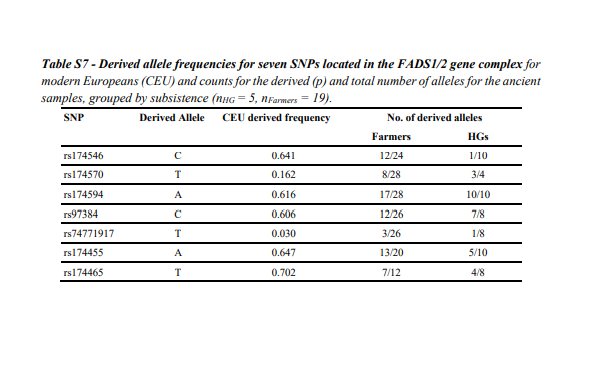
We used the HirisPlexS webtool181,182 to predict pigmentation phenotypes of hair, skin and eyes
for each of the newly sequenced individuals.

In case of missing data in the merged VCF for any of the 38 SNPs used by HirisPlexS, i.e. sites
with low or no depth, the positions were looked up in the BAM files directly (see Table S6). In
BAMs, only sites at least 3 bases away from either end of the read, with a base quality ≥ 25, and
no C/T and G/A SNPs to avoid any effect of PMD on the prediction were considered. In order to
deal with the uncertainty associated to observing alleles in BAM files directly, two HirisPlex input
files were created for each individual: one in which all sites missing from the VCF and meeting
the criteria above were assumed to be homozygous for the allele found in the BAM, and another
in which all positions were assumed to be heterozygous. Running HirisPlexS twice for each sample
resulted in ranges of probabilities for each phenotype (see Table S6). A prediction was accepted
without further explanation if in both runs the same phenotype showed a probability ≥ 0.7182. If
predictions differed between runs, the most parsimonious phenotype was chosen, following the
approach of Walsh in 146. This approach allowed to predict pigmentation phenotypes for all
individuals, except for eye-color in VLASA32 for which no allele at the SNP rs12913832 could
be retrieved.
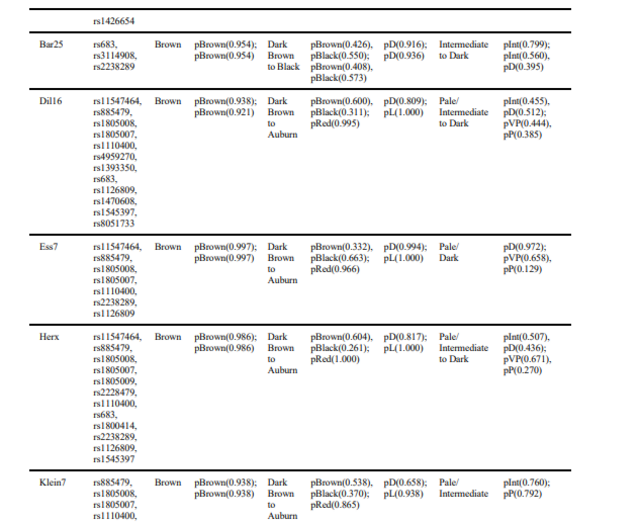
For some individuals, one or several SNPs in the MC1R gene associated with hair color were
missing in the VCF. As described above, we therefore considered both homozygous and
heterozygous states, resulting in predictions of red hair in some cases. Given the overall low
frequency of the derived alleles associated with light pigmentation phenotypes in European
populations183,184 as well as in ancient data78, a red haired phenotype is highly unlikely and we
considered as an artifact. We followed a similar reasoning for skin pigmentation when MC1R
SNPs had to be looked up in the BAM files, as variation in MC1R can also affect the pigmentation
level of the skin185,186. We found that the vast majority of Early Neolithic individuals in our dataset
most likely had an intermediate to light skin complexion, while the two Mesolithic individuals
were inferred to have darker skin tone in comparison. A dark (brown to black) hair color was
inferred for all but two individuals: for LEPE52 and VC3-2, a light brown phenotype was more
likely. Eye color variation was similarly low, with the majority of individuals showing highest
probabilities for brown eyes, except STAR1 and VC3-2, which are inferred to have been blueeyed.
Interestingly, the highest phenotypic variation in our dataset seems to originate from Serbian
individuals.
Pigmentation
We used the HirisPlexS webtool181,182 to predict pigmentation phenotypes of hair, skin and eyes
for each of the newly sequenced individuals.

In case of missing data in the merged VCF for any of the 38 SNPs used by HirisPlexS, i.e. sites
with low or no depth, the positions were looked up in the BAM files directly (see Table S6). In
BAMs, only sites at least 3 bases away from either end of the read, with a base quality ≥ 25, and
no C/T and G/A SNPs to avoid any effect of PMD on the prediction were considered. In order to
deal with the uncertainty associated to observing alleles in BAM files directly, two HirisPlex input
files were created for each individual: one in which all sites missing from the VCF and meeting
the criteria above were assumed to be homozygous for the allele found in the BAM, and another
in which all positions were assumed to be heterozygous. Running HirisPlexS twice for each sample
resulted in ranges of probabilities for each phenotype (see Table S6). A prediction was accepted
without further explanation if in both runs the same phenotype showed a probability ≥ 0.7182. If
predictions differed between runs, the most parsimonious phenotype was chosen, following the
approach of Walsh in 146. This approach allowed to predict pigmentation phenotypes for all
individuals, except for eye-color in VLASA32 for which no allele at the SNP rs12913832 could
be retrieved.

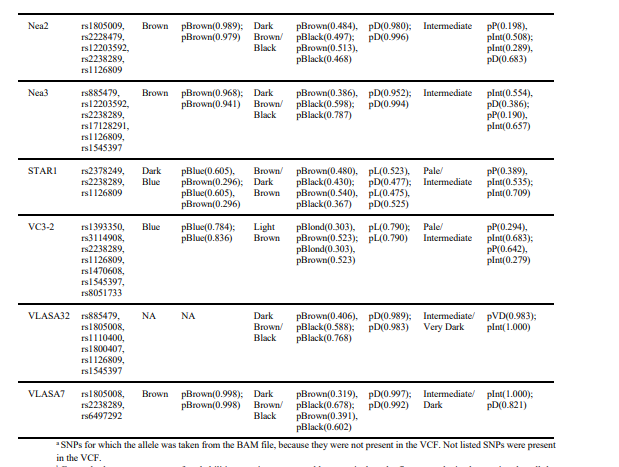
For some individuals, one or several SNPs in the MC1R gene associated with hair color were
missing in the VCF. As described above, we therefore considered both homozygous and
heterozygous states, resulting in predictions of red hair in some cases. Given the overall low
frequency of the derived alleles associated with light pigmentation phenotypes in European
populations183,184 as well as in ancient data78, a red haired phenotype is highly unlikely and we
considered as an artifact. We followed a similar reasoning for skin pigmentation when MC1R
SNPs had to be looked up in the BAM files, as variation in MC1R can also affect the pigmentation
level of the skin185,186. We found that the vast majority of Early Neolithic individuals in our dataset
most likely had an intermediate to light skin complexion, while the two Mesolithic individuals
were inferred to have darker skin tone in comparison. A dark (brown to black) hair color was
inferred for all but two individuals: for LEPE52 and VC3-2, a light brown phenotype was more
likely. Eye color variation was similarly low, with the majority of individuals showing highest
probabilities for brown eyes, except STAR1 and VC3-2, which are inferred to have been blueeyed.
Interestingly, the highest phenotypic variation in our dataset seems to originate from Serbian
individuals.