|
|
Post by Admin on Jun 24, 2016 22:46:45 GMT
 Anatolian farmers first emerged in history in the Mesolithic era and they subsequently expanded in all directions from Europe and South Asia. The Göbekli Tepe site may be associated with Chalcolithic Anatolians who belonged to haplogroup L1a (Lazaridis et al. 2016). L1a, which is common in India and Pakistan (15%), also appears at low frequencies (2-4%) in the Northern part of the Middle East, where the Göbekli Tepe site is located. "The genetic structure of the world’s first farmers" was just published online last week. It concludes that the early farmers of mainland Europe were descended from a population related to Neolithic northwestern Anatolians, which is consistent with an Anatolian origin of farming in Europe.  Extreme regional differentiation in the ancient Near East PCA on present-day West Eurasian populations (Methods) (Extended Data Fig. 1) on which we projected the ancient individuals (Fig. 1b) replicates previous findings of a Europe-Near East contrast along the horizontal Principal Component 1 (PC1) and parallel clines (PC2) in both Europe and the Near East (Extended Data Fig. 1)7,8,13. Ancient samples from the Levant project at one end of the Near Eastern cline, and ancient samples from Iran at the other. The two Caucasus Hunter Gatherers (CHG)9 are less extreme along PC1 than the Mesolithic and Neolithic individuals from Iran, while individuals from Chalcolithic Anatolia, Iran, and Armenia, and Bronze Age Armenia occupy intermediate positions. Qualitatively, the PCA has the appearance of a quadrangle whose four corners are some of the oldest samples: bottom-left: Western Hunter Gatherers (WHG), top-left: Eastern Hunter Gatherers (EHG), bottom-right: Neolithic Levant and Natufians, top-right: Neolithic Iran.  Our data document continuity across the hunter-gatherer / farming transition, separately in the southern Levant and in the southern Caucasus-Iran highlands. The qualitative evidence for this is that PCA, ADMIXTURE, and outgroup f3 analysis cluster Levantine hunter-gatherers (Natufians) with Levantine farmers, and Iranian and Caucasus Hunter Gatherers with Iranian farmers (Fig. 1b; Extended Data Fig. 1; Extended Data Fig. 2). We confirm this in the Levant by showing that its early farmers share significantly more alleles with Natufians than with the early farmers of Iran: the statistic f4(Levant_N, Chimp; Natufian, Iran_N) is significantly positive (Z=13.6). The early farmers of the Caucasus-Iran highlands similarly share significantly more alleles with the hunter-gatherers of this region than with the early farmers from the Levant: the statistic f4(Iran_N, Chimp; Caucasus or Iran highland hunter-gatherers, Levant_N) is significantly positive (Z>6).  Among first farmers, those of the Levant trace ~2/3 of their ancestry to people related to Natufian hunter-gatherers and ~1/3 to people related to Anatolian farmers (Supplementary Information, section 7). Western Iranian first farmers cluster with the likely Mesolithic HotuIIIb individual and more remotely with hunter-gatherers from the southern Caucasus (Fig. 1b), and share alleles at an equal rate with Anatolian and Levantine early farmers (Supplementary Information, section 7), highlighting the long-term isolation of western Iran.  During subsequent millennia, the early farmer populations of the Near East expanded in all directions and mixed, as we can only model populations of the Chalcolithic and subsequent Bronze Age as having ancestry from two or more sources. The Chalcolithic people of western Iran can be modelled as a mixture of the Neolithic people of western Iran, the Levant, and Caucasus Hunter Gatherers (CHG), consistent with their position in the PCA (Fig. 1b). Admixture from populations related to the Chalcolithic people of western Iran had a wide impact, consistent with contributing ~44% of the ancestry of Levantine Bronze Age populations in the south and ~33% of the ancestry of the Chalcolithic northwest Anatolians in the west. Our analysis show that the ancient populations of the Chalcolithic Iran, Chalcolithic Armenia, Bronze Age Armenia and Chalcolithic Anatolia were all composed of the same ancestral components, albeit in slightly different proportions (Fig. 4b; Supplementary Information, section 7). 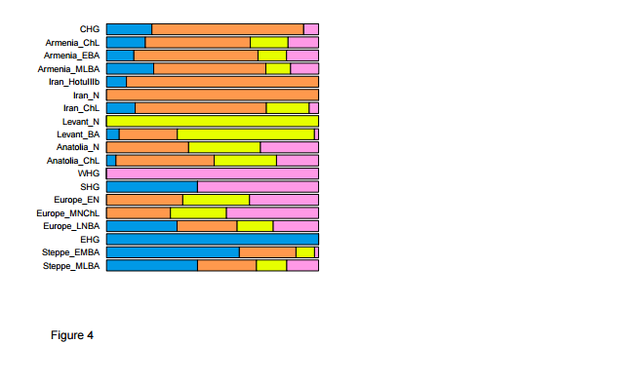 Admixture did not only occur within the Near East but extended towards Europe. To the north, a population related to people of the Iran Chalcolithic contributed ~43% of the ancestry of early Bronze Age populations of the steppe. The spread of Near Eastern ancestry into the Eurasian steppe was previously inferred7 268 without access to ancient samples, by hypothesizing a population related to present-day Armenians as a source7,8. To the west, the early farmers of mainland Europe were descended from a population related to Neolithic northwestern Anatolians8. This is consistent with an Anatolian origin of farming in Europe but does not reject other sources, since the spatial distribution of the Anatolian/European-like farmer populations is unknown. We can rule out the hypothesis that European farmers stem directly from a population related to the ancient farmers of the southern Levant30,31.  In South Asia, our dataset provides insight into the sources of Ancestral North Indians (ANI), a West Eurasian related population that no longer exists in unmixed form but contributes a variable amount of the ancestry of South Asians35,36 (Supplementary Information, section 9) (Extended Data Fig. 4). We show that it is impossible to model the ANI as being derived from any single ancient population in our dataset. However, it can be modelled as a mix of ancestry related to both early farmers of western Iran and to people of the Bronze Age Eurasian steppe; all sampled South Asian groups are inferred to have significant amounts of both ancestral types. The demographic impact of steppe related populations on South Asia was substantial, as the Mala, a south Indian population with minimal ANI along the ‘Indian Cline’ of such ancestry35,36 is inferred to have ~18% steppe-related ancestry, while the Kalash of Pakistan are inferred to have ~50%, similar to present-day northern Europeans7. 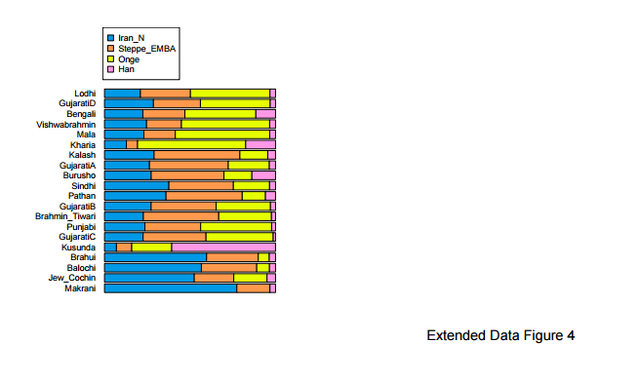 Applying this while withholding major populations, we validated some of our key inferences, successfully inferring mixture proportions consistent with those obtained when the populations are included in the analysis. Application of this methods highlights the impact of Ancient North Eurasian (ANE) ancestry related to the ~22,000 BCE Mal’ta 1 and ~15,000 BCE Afontova Gora 215 311 on populations living in Europe, the Americas, and Eastern Eurasia. Eastern Eurasians can be modelled as arrayed along a cline with different proportions of ANE ancestry (Supplementary Information, section 11; Extended Data Fig. 7), ranging from ~40% ANE in Native Americans matching previous findings13,15, to no less than ~5-10% ANE in diverse East 316 Asian groups including Han Chinese (Extended Data Fig. 4; Extended Data Fig. 6f). We also document a cline of ANE ancestry across the east-west extent of Eurasia. Eastern Hunter Gatherers (EHG) derive ~3/4 of their ancestry from the ANE (Supplementary Information,section 11); Scandinavian hunter-gatherers7,8,13 319 (SHG) are a mix of EHG and WHG; and WHG are a mix of EHG and the Upper Paleolithic Bichon from Switzerland (Supplementary Information, section 7). Northwest Anatolians—with ancestry from a population related to European hunter-gatherers (Supplementary Information, section 7)—are better modelled if this ancestry is taken as more extreme than Bichon (Supplementary Information, section 10). doi: dx.doi.org/10.1101/059311 |
|
|
|
Post by Admin on Jun 25, 2016 22:41:56 GMT
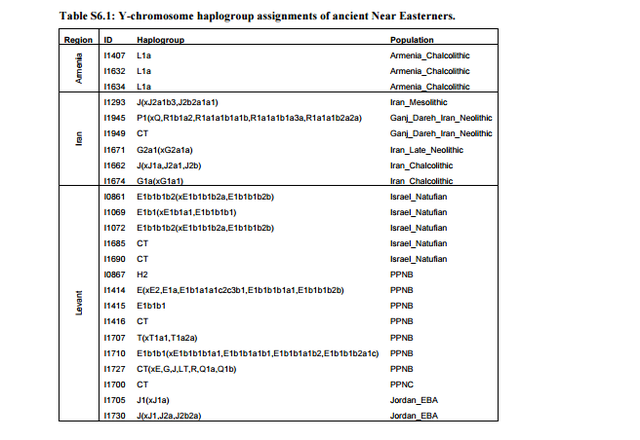 We highlight two aspects of the Y-chromosome distribution in the ancient Near East. First, the Mesolithic individual from Iran belonged to haplogroup J. This has also been detected in two hunter-gatherers from the Upper Paleolithic in Georgia11, as well as in a hunter-gatherer from Karelia in northwest Russia5, suggesting that it had a widespread early distribution prior to the spread of farming with which its current distribution was initially associated12. While the hunter-gatherers from Georgia resemble the one from Iran (Fig. 1b), their whole genome data shows very different patterns from the Eastern European hunter-gatherer who also possessed this haplogroup, as well as from a singleton Anatolian early farmer who belonged to haplogroup J2a5 . This should serve as a note of caution against the idea that Y-chromosome lineages can be thought as markers of populations and population movements: the two sometimes coincide as with the sudden increase of haplogroups R1a and R1b in mainland Europe coinciding with an influx of steppe migrants around ~4,500 years ago6, or the spread of migrants carrying haplogroup G2a2 during the early Neolithic settlement of Europe5 . With this caveat in mind, it is nonetheless interesting that haplogroup J was present in EHG, CHG, and Mesolithic Iran, given the evidence for a relationship between these populations presented in Supplementary Information, sections 4, 7. Additional sampling of earlier individuals from eastern Europe and western Asia may elucidate the nature of this relationship. We also note one instance of a possible relationship between haplogroup J and the appearance of possible admixture event: two of the Bronze Age samples from the Levant belong to this haplogroup, while none of the earlier samples of Natufians and Pre-Pottery Neolithic individuals do. In Supplementary Information section 7, we infer that Levant_BA can be modeled as a mixture of Levant_N and Iran_ChL. The Y chromosome results suggest that haplogroup J migrants from the north of the Levant may have been involved in this mixture event.  Second, we observed that all three Natufian individuals that could be assigned to a specific haplogroup belonged to haplogroup E1b1. This is thought to have an East African origin, and a 4,500-year old individual from the Ethiopian highlands13 belonged to it. An African origin of the Natufians has been proposed based on their ‘Sub-Saharan’ cranial morphology14 in comparison to later West Eurasian samples. However, when we test whether present-day Sub-Saharan Africans share more alleles with Natufians than they do with other ancient West Eurasian individuals, we do not find the expected asymmetry if Natufians have ancestry related to present-day Sub-Saharan Africans (Extended Data Table 1).  The existence of haplogroup E1b1 in ancient East Africa and the nearby Levant in the two earliest samples from both regions raises questions about its ultimate origin. This haplogroup continues to exist in the Pre-Pottery Neolithic period, but it is not documented in ancient Europe or the ancient Near East outside the Levant. This appears to be consistent with E1b1 being intrusive to the Near East from nearby Africa, rather than its having a long-term presence in the region.  An association of E1b1 migrants from Africa with Basal Eurasian ancestry is possible (Basal Eurasians are so named because of their phylogenetic position basal to other Eurasians not for their geographical provenance; African migrants that did not participate in the initial Out-ofAfrica expansion would occupy such a basal position to other Eurasians). However, such ancestry is not limited to the Levant, but also extends to the whole of Near East (where E1b1 chromosomes have not been detected). Thus, we think that both the late entry scenario of Basal Eurasian ancestry into the Near East (associated with gene flow from Africa), or its earlier presence15 in anatomically modern-humans from the Levant and Arabia >100,000 years 16,17 are still plausible. bioRxiv preprint first posted online Jun. 16, 2016; doi: dx.doi.org/10.1101/059311. |
|
|
|
Post by Admin on Jun 27, 2016 22:47:31 GMT
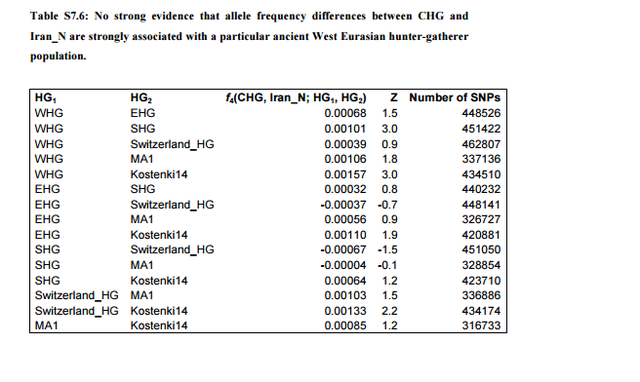 Caucasus hunter-gatherers can be modeled as a mixture of Neolithic Europe and European hunter-gatherers The Caucasus hunter-gatherers cluster with the Iran_N in PCA (Fig. 1b), but are also shifted towards Europe. We verify with f4-statistics that the CHG and Iran_N do not form a clade, but rather that most ancient West Eurasian populations share more alleles with the CHG than with the Iran_N (Fig. S7.6). Some of these statistics could be interpreted in terms of gene flow into Europe, associated with the migration of the Yamnaya that included a component from theCaucasus1, for which the CHG could be a representative16. However, similar to the argument we previously made for the relationship of the Levantine Neolithic to Natufians, the positive f4-statistics that involve European hunter-gatherers or even MA1 cannot be explained in this manner (as they predate the Yamnaya by thousands of years). 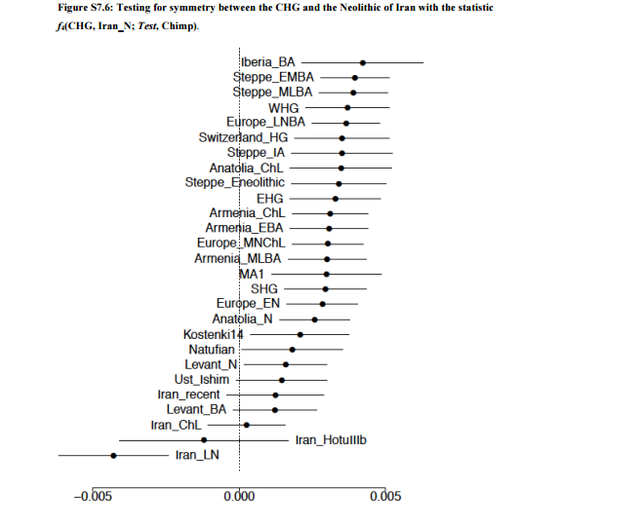 An alternative explanation is that the excess of genetic drift shared by the CHG and European hunter-gatherers is caused by the Iran_N having more Basal Eurasian ancestry than the CHG.The statistic f4(CHG, Iran_N; Ust_Ishim, Chimp) = 0.00145 (Z=2.8; 762,778 SNPs) is suggestive that this is the case. The same could be the case for the Iran_HotuIIIb population,which is similar in magnitude, but on a much lower number of SNPs: f4(Iran_HotuIIIb, Iran_N; Ust_Ishim, Chimp) = 0.00154 (Z=1.6; 116,123 SNPs). 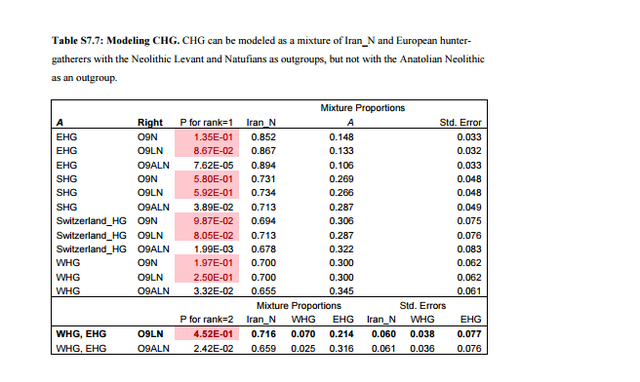 There does not appear to be any evidence that the difference between CHG and Iran_N is strongly correlated with any particular European hunter-gatherer population (Table S7.6), with the only two comparisons that reach significant (|Z|=3) hinting that CHG shares more alleles with WHG than with SHG or Kostenki14. We can model CHG as a mixture of Iran_N and different European hunter-gatherer populations (Table S7.7), with an estimate of 71.6±6.0% Iran_N, 7.0±3.8% WHG, 21.4±7.7% EHG. A sanity check of this estimate is that it predicts 0.716*.482=0.345 Basal Eurasian ancestry in the CHG, virtually identical to the 0.347±0.058 of Supplementary Information, section 4. |
|
|
|
Post by Admin on Jul 26, 2016 21:41:44 GMT
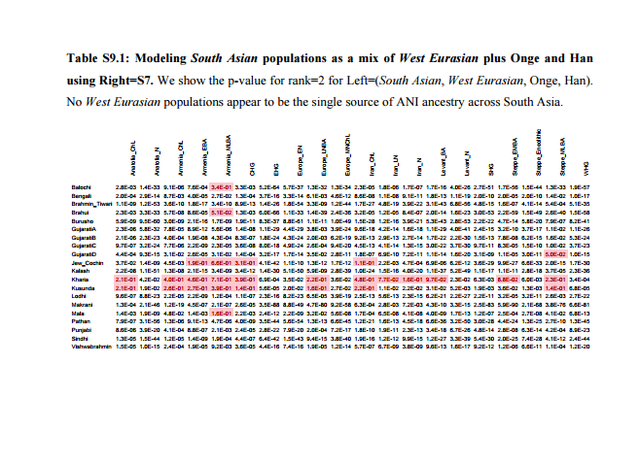 Previous studies1,2 have uncovered evidence of admixture in South Asian populations from an “Ancestral North Indian” (ANI) source that is related to West Eurasian populations. It has been proposed that populations of the Caucasus such as Georgians were related to the ANI1,3, a claim that has found additional support by the analysis of the Caucasus hunter-gatherers (CHG) from Georgia4 which appear to be a source of ancestry for South Asian populations. However, South Asia is also linked to the Eurasian steppe5 by the analysis of Y-chromosomes which detected the presence of Ychromosome haplogroup R1a1a1b2-Z93 as a common element in ancient Bronze Age populations of eastern Europe5, Mongolia6, and Central7/South Asia8 and which may mark spread of Indo-European languages eastward as suggested by the steppe origin theory of Indo-European languages9.  The admixture history of ANI into the Indian subcontinent is likely to be complex, as there is evidence of more than one layer of admixture within the last 4,000 years1. Moreover, unlike Europe where a substantial number of pre-agricultural hunter-gatherers is available for study4,5,10-16, the earliest population substratum of the “Ancestral South Indians” (ASI) is only indirectly known by its distant relationship2 to the Onge hunter-gatherers from the Andaman Islands17, a population that may be an imperfect proxy for the ASI. There is also evidence that Indian populations have ancestry related to Austroasiatic and Tibeto-Burman groups2,18, although many of them can be modeled as a simpler mixture involving only the ANI-ASI ancestral populations1,2. 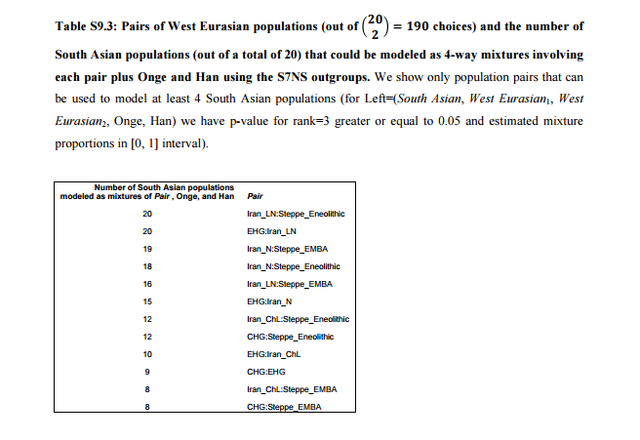 In Table S9.3 we see that only 12 pairs of West Eurasian populations can be used to model at least 4 South Asian populations. All these pairs involve a steppe population and a population from Iran or CHG. Only steppe/Iran combinations can model all or nearly all South Asian populations successfully. In Table S9.4 we list the inferred mixture proportions for the top 3 pairs that can be used to model nearly all South Asian populations in our dataset. Ancestry from both Iran and Steppe-related sources is pervasive across South Asia. Estimated proportions of EHG ancestry are lower than Steppe_Eneolithic ancestry which are lower than Steppe_EMBA ancestry, consistent with the dilution of EHG ancestry from a southern source5 related to populations of Iran (Supplementary Information, section 7), thus more Steppe ancestry is required when the Steppe source has less EHG ancestry. 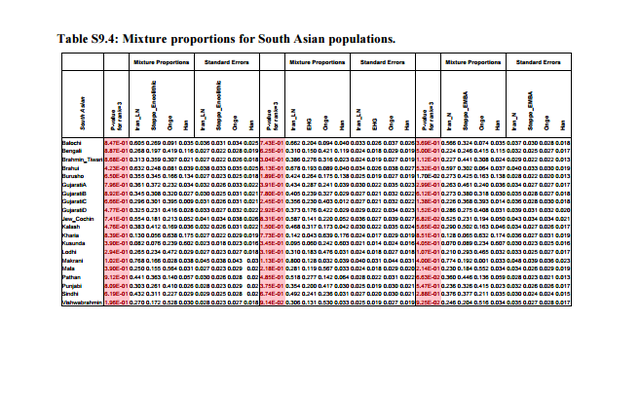 In Extended Data Fig. 4 we plot mixture proportions using Iran_N and Steppe_EMBA as sources as these source populations have the greatest sample sizes. Nonetheless, we do not claim that any particular pair of populations is directly ancestry to South Asian populations. This section places constraints on the possible sources of ANI ancestry in India by showing that single populations of ancient West Eurasia are not feasible sources (Table S9.1) while pairs of populations involving Steppe/Iran ancestry are (Tables S9.2 and S9.3). The geographical interpretation of this finding is unclear; mixtures of Steppe/Iran-related ancestry (such as Steppe_EMBA and Steppe_Eneolithic) were formed in West Eurasia and it is possible that a currently unsampled such mixed population is responsible for the ANI ancestry in South Asia. Alternatively, admixture may have taken place by combinations of Steppe/Iran-related groups in central or south Asia. The analysis in this section reconciles the evidence presented in the first paragraph regarding the origin of the ANI by showing that is may be related both to “southern” populations related to Iran and the Caucasus and to “northern” steppe populations. Our results do not resolve the relationship between ANI and the origin of Indo-European speakers in South Asia, in the sense that they reveal that South Asian populations have ancestry both from regions related to the Eurasian steppe and ancient Iran, which is compatible with alternative homeland solutions12. West Eurasian-related ANI ancestry in South Asia may pre-date, coincide with, or postdate Indo-European dispersals, although a partial link between the two is suggested by the evidence for Bronze Age admixture in India1 that contributed a large portion of ancestry especially in Indo-European speakers1,2 whose magnitude would be compatible with major linguistic change. However, ANI ancestry related to both ancient Iran and the steppe is found across South Asia (Table S9.4) making it difficult to associate it strongly with any particular language family (Indo-European or otherwise).  Nonetheless, the fact that we can reject West Eurasian population sources from Anatolia, mainland Europe, and the Levant diminish the likelihood that these areas were sources of Indo-European (or other) languages in South Asia. While the Early/Middle Bronze Age ‘Yamnaya’-related group (Steppe_EMBA) is a good genetic match (together with Neolithic Iran) for ANI, the later Middle/Late Bronze Age steppe population (Steppe_MLBA) is not. Steppe_MLBA includes Sintashta and Andronovo populations who have been proposed as identical to or related to ancestral Indo-Iranians9,19, as well as the Srubnaya from eastern Europe which are related to South Asians by their possession of Y-chromosome haplogroup R1a1a1b2-Z935. A useful direction of future research is a more comprehensive sampling of ancient DNA from steppe populations, as well as populations of central Asia (east of Iran and south of the steppe), which may reveal more proximate sources of the ANI than the ones considered here, and of South Asia to determine the trajectory of population change in the area directly. |
|
|
|
Post by Admin on Jul 31, 2016 21:26:46 GMT
Neanderthal ancestry All statistics presented in this section are computed with 'qpF4ratio' from the ADMIXTOOLS package (6). We implement the f4-ratio scheme described by Fu et al., 2016(14), in section 3 (pp. 21) of the SM, however, we use reference population data from Lazaridis et al., 2014 (82) instead of full genome sequences from the Simons Genome Diversity Project Dataset (104). Therefore, all our results are based on a maximum of around 500K SNPs, which explains the differences we observe compared to the original values for samples published in Fu et al. that we re-analyse here. 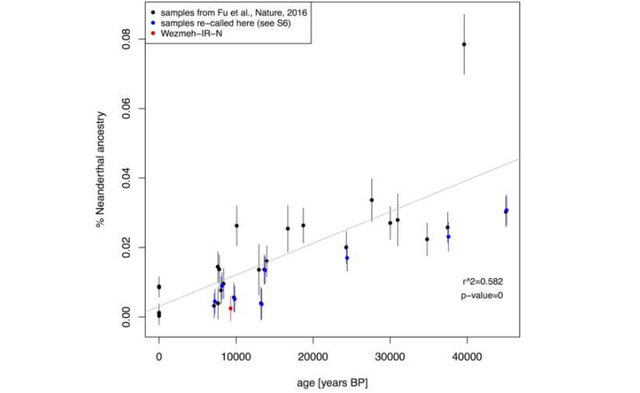 The Neanderthal ancestry proportions are given in Table S21. Samples from three different sources are included (specified in column ‘dataset’): ‘Fu’, ‘Lazaridis’, and ‘PAT_gt’. As already mentioned above, we re-analyzed the samples published in Fu et al. 2016 (labeled ‘Fu’), excluding low coverage samples as in Figure 2 of Fu et al.. This is ‘pseudo-haploid’ data resulting from a random allele calling procedure, except for Loschbour, Ust_’Ishim and Stuttgart, which are diploid. Contemporary populations are from Lazaridis et al., 2014 (82) (labeled ‘Lazaridis’), and consist of diploid data. Finally, samples labeled ‘PAT_gt’ are samples we called with our genotype caller described in Supplementary section S6.  Decrease of Neanderthal ancestry over time. We see the same trend of reduction in Neanderthal ancestry with time as observed by Fu et al. 2016 (Fig. S22). We obtain good agreement of inferred ancestry proportions between samples that are both in the original Fu et al. dataset and were re-called with our genotype caller, including the ‘pseudo-haploid’ Fu et al . samples without heterozygote sites. We infer very low levels of Neanderthal ancestry for WC1. However, WC1 does not behave as an outlier as temporally close samples are inferred to have similar proportions. Science 14 Jul 2016: DOI: 10.1126/science.aaf7943 |
|






















