|
|
Post by Admin on Jul 23, 2016 21:33:53 GMT
R1a-M420 is one of the most widely spread Y-chromosome haplogroups; however, its substructure within Europe and Asia has remained poorly characterized. Using a panel of 16 244 male subjects from 126 populations sampled across Eurasia, we identified 2923 R1a-M420 Y-chromosomes and analyzed them to a highly granular phylogeographic resolution. Whole Y-chromosome sequence analysis of eight R1a and five R1b individuals suggests a divergence time of ~25 000 (95% CI: 21 300–29 000) years ago and a coalescence time within R1a-M417 of ~5800 (95% CI: 4800–6800) years. The spatial frequency distributions of R1a sub-haplogroups conclusively indicate two major groups, one found primarily in Europe and the other confined to Central and South Asia. Beyond the major European versus Asian dichotomy, we describe several younger sub-haplogroups. Based on spatial distributions and diversity patterns within the R1a-M420 clade, particularly rare basal branches detected primarily within Iran and eastern Turkey, we conclude that the initial episodes of haplogroup R1a diversification likely occurred in the vicinity of present-day Iran.  More than 10% of men in a region extending from South Asia to Scandinavia share a common ancestor in hg R1a-M420, and the vast majority fall within the R1a1-M17/M198 subclade.22 Although the phylogeography of R1b-M343 has been described, especially in Western and Central Europe,15, 23, 24, 25 R1a1 has remained poorly characterized. Previous work has been limited to a European-specific subgroup defined by the single-nucleotide polymorphism (SNP) called M458.22, 26, 27, 28, 29, 30 However, with the discovery of the Z280 and Z93 substitutions within Phase 1 1000 Genomes Project data1 and subsequent genotyping of these SNPs in ~200 samples, a schism between European and Asian R1a chromosomes has emerged.31 We have evaluated this division in a larger panel of populations, estimated the split time, and mapped the distributions of downstream sub-hgs within seven regions: Western/Northern Europe, Eastern Europe, Central/South Europe, the Near/Middle East, the Caucasus, South Asia, and Central Asia/Southern Siberia. Subgroups of both R1a-Z282 and R1a-Z93 exhibit geographic localization within the broad distribution zone of R1a-M417/Page7. Among R1a-Z282 subgroups (Figure 2), the highest frequencies (~20%) of paragroup R1a-Z282* chromosomes occur in northern Ukraine, Belarus, and Russia (Figure 2b). The R1a-Z284 subgroup (Figure 2c) is confined to Northwest Europe and peaks at ~20% in Norway, where the majority of R1a chromosomes (24/26) belong to this clade. We found R1a-Z284 to be extremely rare outside Scandinavia. R1a-M458 (Figure 2d) and R1a-M558 (Figure 2e) have similar distributions, with the highest frequencies observed in Central and Eastern Europe. R1a-M458 exceeds 20% in the Czech Republic, Slovakia, Poland, and Western Belarus. The lineage averages 11–15% across Russia and Ukraine and occurs at 7% or less elsewhere (Figure 2d). Unlike hg R1a-M458, the R1a-M558 clade is also common in the Volga-Uralic populations. R1a-M558 occurs at 10–33% in parts of Russia, exceeds 26% in Poland and Western Belarus, and varies between 10 and 23% in the Ukraine, whereas it drops 10-fold lower in Western Europe. In general, both R1a-M458 and R1a-M558 occur at low but informative frequencies in Balkan populations with known Slavonic heritage. The rarity of R1a-M458 and R1a-M558 among Central Asian and South Siberian R1a samples (4/301; Supplementary Table 4) suggests low levels of historic Slavic gene flow. In the complementary R1a-Z93 haplogroup, the paragroup R1a-Z93* (Figure 3b) is most common (>30%) in the South Siberian Altai region of Russia, but it also occurs in Kyrgyzstan (6%) and in all Iranian populations (1–8%). R1a-Z2125 (Figure 3c) occurs at highest frequencies in Kyrgyzstan and in Afghan Pashtuns (>40%). We also observed it at greater than 10% frequency in other Afghan ethnic groups and in some populations in the Caucasus and Iran. Notably, R1a-M780 (Figure 3d) occurs at high frequency in South Asia: India, Pakistan, Afghanistan, and the Himalayas. The group also occurs at >3% in some Iranian populations and is present at >30% in Roma from Croatia and Hungary, consistent with previous studies reporting the presence of R1a-Z93 in Roma.31, 51 Finally, the rare R1a-M560 was only observed in four samples: two Burushaski speakers from north Pakistan, one Hazara from Afghanistan, and one Iranian Azeri. Origin of hg R1a To infer the geographic origin of hg R1a-M420, we identified populations harboring at least one of the two most basal haplogroups and possessing high haplogroup diversity. Among the 120 populations with sample sizes of at least 50 individuals and with at least 10% occurrence of R1a, just 6 met these criteria, and 5 of these 6 populations reside in modern-day Iran. Haplogroup diversities among the six populations ranged from 0.78 to 0.86 (Supplementary Table 4). Of the 24 R1a-M420*(xSRY10831.2) chromosomes in our data set, 18 were sampled in Iran and 3 were from eastern Turkey. Similarly, five of the six observed R1a1-SRY10831.2*(xM417/Page7) chromosomes were also from Iran, with the sixth occurring in a Kabardin individual from the Caucasus. Owing to the prevalence of basal lineages and the high levels of haplogroup diversities in the region, we find a compelling case for the Middle East, possibly near present-day Iran, as the geographic origin of hg R1a. Spatial dynamics of R1a lineage frequencies We conducted a spatial autocorrelation analysis of the two primary subgroups of R1a (Z282 and Z93) and of each of their subgroups independently (Supplementary Figure 3). Each correlogram was statistically significant. We observed clinal distributions (continually decreasing frequency with increasing geographic distance) across a large geographic area in the two macrogroups and in M558 and M780 as well. One group (Z2125) did not reveal any discernible pattern, and the analysis of four groups (Z282*, Z284, M458, and Z93*) indicated potential clinal distributions that do not extend across the full geographic range under study. Therefore, we also analyzed partial ranges for Z282* and M458 in Europe, the Caucasus, and the Middle East, and for Z284 in Europe, but these partial range analyses also failed to yield evidence of clinal distributions. We also conducted PCA of R1a subgroups (Figure 4). The first principal component explains 21% of the variation and separates European populations at one extreme from those of South Asia at the other. The second explains 14.7% of the variation and is driven almost exclusively by the high presence of M582 among some Jewish populations, particularly the Ashkenazi Jews. PC2 separates them from all other populations. When we consider haplogroups rather than populations (Supplementary Figure 4), we see that the clustering of European populations is due to their high frequencies of M558, M458, and Z282*, whereas the M780 and Z2125* lineages account for the South Asian character of the other extreme. To put our frequency distribution maps, PCA analyses, and autocorrelation results in archaeological context, we note that the earliest R1a lineages (genotyped at just SRY10381.2) found thus far in European ancient DNA date to 4600 years before present (YBP), a time corresponding to the Corded Ware Culture,53 whereas three DNA sample extracts from the earlier Neolithic Linear Pottery Culture (7500–6500 YBP) period were reported as G2a-P15 and F-M89(xP-M45) lineages.54 This raises the possibility of a wide and rapid spread of R1a-Z282-related lineages being associated with prevalent Copper and Early Bronze Age societies that ranged from the Rhine River in the west to the Volga River in the east55 including the Bronze Age Proto-Slavic culture that arose in Central Europe near the Vistula River.56 It may have been in this cultural context that hg R1a-Z282 diversified in Central and Eastern Europe. The corresponding diversification in the Middle East and South Asia is more obscure. However, early urbanization within the Indus Valley also occurred at this time57 and the geographic distribution of R1a-M780 (Figure 3d) may reflect this. European Journal of Human Genetics (2015) 23, 124–131; doi:10.1038/ejhg.2014.50; published online 26 March 2014 |
|
|
|
Post by Admin on Nov 2, 2017 19:06:32 GMT
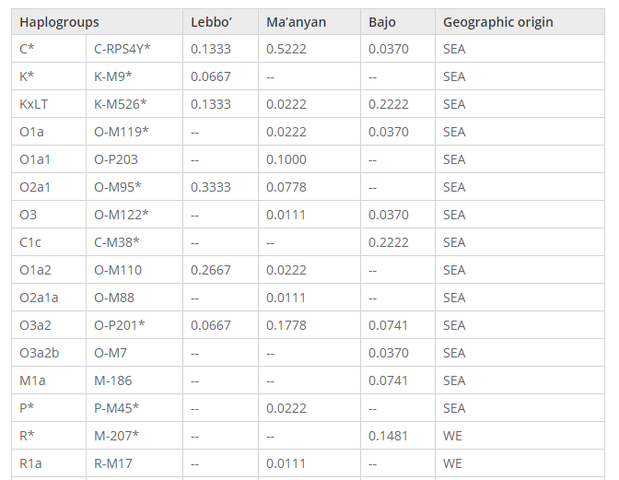 Table 1 Y chromosome haplogroup frequencies in the Ma’anyan, Lebbo’ and Bajo Y chromosome and mitochondrial DNA classification Based on analysis of 96 Y chromosome binary markers (Additional file 1: Table S1), the majority of men in the Ma’anyan, Lebbo’ and Bajo carry haplogroups previously found in Southeast Asia, particularly C*, K*, and O* (Table 1). Only a few individuals carry Y chromosomes belonging to Western Eurasian haplogroups: R* (M207) [40,41] was found in four Bajo individuals, R1a (M17) [42,43] was found in one Ma’anyan individual, while the western Eurasian haplogroups L1a (M76) and T1a (M70) [44,45] were found in one and two Bajo individuals, respectively. Indian haplogroup R*, which includes R1a, has previously been identified in Bali, Java, Borneo, and Mandar (Additional file 2: Table S2), and thus could conceivably have transited through Indonesia (as opposed to a direct connection), but T1a and L1a have not been identified to date in any Indonesian population. On the mitochondrial DNA (Table 2), the frequency distributions of haplogroups found in the Ma’anyan, Lebbo’ and Bajo are broadly similar to, and consistent with, patterns of maternal lineages in Indonesia. Indeed, four main geographical/historical affiliations can be observed: mainland Asia, the Austronesian expansion, western Eurasia/India, and New Guinea. In brief, mainland Asian mtDNA haplogroups (such as B4c2, M73, M74, M12) are carried by a majority of individuals (64%), followed by haplogroups that have been putatively linked with an Austronesian expansion out of Taiwan (such as B4a1a1, M7c1a4, F1*, E*; 32%). The remaining lineages likely derive from India and west Eurasia, and were only observed among the Ma’anyan (M2, M5a4, and M35a, ranging in frequency from 0.6-1.9%). The presence of Indian and other western Eurasian genetic traces has been observed previously in Borneo, as well as Sumatra, Java and Bali [46,47] (Additional file 3: Table S3). Indian haplogroups are restricted to western Indonesia, particularly in regions historically involved in the ancient trading networks of the Hindu kingdoms (such as Srivijaya and Majapahit). Among the Bajo, we also observed the M1a Y haplogroup and Q1 mitochondrial haplogroup, which likely traces its ancestry to New Guinea or eastern Indonesia [39,48-50]. These haplogroups represent a trace of Papuan genetic input. This is perhaps due to the extensive trading network of the Bajo eastward to New Guinea [30] and/or earlier westward expansions of Papuan speakers from New Guinea to eastern Indonesia [51].  Paternal lineage proximity to Malagasy Shared lineages Among the haplogroups shared between Malagasy and Indonesians (Additional file 4: Table S4), four originated in Island Southeast Asia (C, O1a, O1a2, O2a1*), while six have western Eurasian origins (J1, J2, J2b, T*, L* and R1a). The Ma’anyan and five other Indonesian groups, all located around the Sulawesi sea (east Kalimantan Dayak, Java, Bali, Mandar and Sumba), share four of these Island Southeast Asian haplogroups. Importantly, Malagasy uniquely share just one subhaplogroup (O2a1a1-M88) with Ma’anyan, and this lineage has not been discovered in other regions of Indonesia. O2a1a1 may therefore be a marker of male genetic contributions from southern Borneo to Madagascar. Shared Y chromosome haplogroups with a west Eurasian origin (J, T, L and R1a) (Additional file 4: Table S4) are also present in Indonesian populations, but only in the south and west of the Sulawesi sea. They occur at low frequency (<0.1%) in Java, Bali, Mandar and Bajo, but R1a is the only west Eurasian haplogroup identified in southern Borneo (Ma’anyan and east Kalimantan Dayak). However, west Eurasian lineages in Indonesia and Madagascar may result from independent dispersal events. Indeed, Indian and Arab traders have been active on both side of the Indian Ocean within the last three and one thousand years, respectively [47,52-56]. Therefore, west Eurasian haplogroups shared between Malagasy and Indonesians may have originated from Indonesia, or alternatively, they may have been obtained directly from southwestern Eurasia (the Middle East or India).  FST values based on Y chromosome haplogroup frequencies (Additional file 5: Table S5) were visualized on a multidimensional scaling (MDS) plot (Figure 1). Due to their statistically supported genetic homogeneity, Malagasy groups were pooled. The MDS plot (Figure 1) shows that Malagasy Y chromosome lineages are an outlier to Indonesian populations, in a similar way to certain Indonesian population outliers (Mentawai, Nias, Besemah, Semende). Y chromosome FST values between Malagasy and Indonesians are relatively high (FST > 0.2; Additional file 5: Table S5), mostly driven by the substantial African component of Malagasy (~65% of the paternal gene pool, but only ~30% of mtDNA [25,24]). No significant differences were observed to suggest specific genetic connections between Malagasy and eastern versus western Indonesians (Mann–Whitney U-test: P = 0.06). The Ma’anyan and other populations from Borneo cluster together with western Indonesian groups, including several population outliers (Mentawai, Nias, Besemah, Semende). The Bajo cluster with eastern Indonesian groups, consistent with the well-documented genetic division between western and eastern Indonesia broadly along the Wallace line. When FST values are visualized with Surfer (Figure 2), the Indonesian populations with closest affinity to Malagasy (FST in the lower quartile of the range) are from regions near Wallace’s line in the west and south of the Sulawesi sea (southern Sulawesi, eastern Borneo and the Lesser Sunda islands). Populations with highest affinity to Malagasy are Mandar (Sulawesi), Flores (Lesser Sunda), Bajo (Sulawesi), and east Kalimantan Dayak and Lebbo’ (Borneo) (Additional file 5: Table S5). These results are supported by a linear optimization method, which aims to find the combination of Indonesian populations that most closely resembles the observed haplogroup diversity in Malagasy. This algorithm highlights two populations from the west and south of the Sulawesi sea, the Mandar (Sulawesi) and Lebbo’ (Borneo), as populations that produce a Y chromosome genetic profile most closely resembling the observed pattern, while still accounting for the predominantly African genetic background found in Malagasy (Additional file 6: Figure S1).  Figure 2 Map of Y chromosome F ST values obtained by pairwise comparison between Malagasy and Indonesian populations. These geographical regions comprised part of the trading sphere of the Srivijaya empire, including several Javanese kingdoms that played a crucial role in the region: Heluodan (5th century), Tarumanagara (5th century), Walaing (Chinese Heling, 7th-8th centuries), Kahuripan/Kediri (11th century), Singasari (13th century) and Majapahit (13th -15th centuries) [57]. This region also hosted several houseboat nomad groups (such as the Bajo), which had ample opportunities to incorporate men from a wider regional watershed. The Ma’anyan from southern Borneo do not show any privileged link with Malagasy (indeed, they have a relatively high FST value showing genetic differentiation), despite being the only Indonesian population that shares Y haplogroup O2a1a with Malagasy. This may indicate that the genetic contribution of Ma’anyan was limited, either due to the recent arrival of this lineage in Ma’anyan, or perhaps O2a1a has since been lost or is still undetected in other Indonesian populations. BMC Genomics201516:191 DOI: 10.1186/s12864-015-1394-7 |
|
|
|
Post by Admin on Dec 22, 2021 18:18:14 GMT
 The phylogenetic and geographic structure of Y-chromosome haplogroup R1a
R1a-M420 is one of the most widely spread Y-chromosome haplogroups; however, its substructure within Europe and Asia has remained poorly characterized. Using a panel of 16 244 male subjects from 126 populations sampled across Eurasia, we identified 2923 R1a-M420 Y-chromosomes and analyzed them to a highly granular phylogeographic resolution. Whole Y-chromosome sequence analysis of eight R1a and five R1b individuals suggests a divergence time of B25 000 (95% CI: 21 300–29 000) years ago and a coalescence time within R1a-M417 of B5800 (95% CI: 4800–6800) years. The spatial frequency distributions of R1a sub-haplogroups conclusively indicate two major groups, one found primarily in Europe and the other confined to Central and South Asia. Beyond the major European versus Asian dichotomy, we describe several younger sub-haplogroups. Based on spatial distributions and diversity patterns within the R1a-M420 clade, particularly rare basal branches detected primarily within Iran and eastern Turkey, we conclude that the initial episodes of haplogroup R1a diversification likely occurred in the vicinity of present-day Iran.
European Journal of Human Genetics (2015) 23, 124–131; doi:10.1038/ejhg.2014.50; published online 26 March 2014 Introduction High-throughput resequencing efforts have uncovered thousands of Y-chromosome variants that have enhanced our understanding of this most informative locus’ phylogeny, both through the resolution of topological ambiguities and by enabling unbiased estimation of branch lengths, which, in turn, permit timing estimates.1, 2, 3, 4, 5 The International Society of Genetic Genealogy10 has aggregated these variants and those discovered with previous technologies into a public resource that population surveys can leverage to further elucidate the geographic origins of and structure within haplogroups.6, 7, 8, 9, 10, 11, 12, 13 Y-chromosome haplogroup R (hg R) is one of 20 that comprise the standardized global phylogeny.14 It consists of two main components: R1-M173 and R2-M47915 (Figure 1). Within R1-M173, most variation extant in Eurasia is confined to R1a-M420 and R1b-M343.16 In Europe, R1a is most frequent in the east, and R1b predominates in the west.17 It has been suggested that this division reflects episodic population expansions during the post-glacial period, including those associated with the establishment of agricultural/pastoral economies.3, 18, 19, 20, 21 |
|
|
|
Post by Admin on Dec 22, 2021 20:24:25 GMT
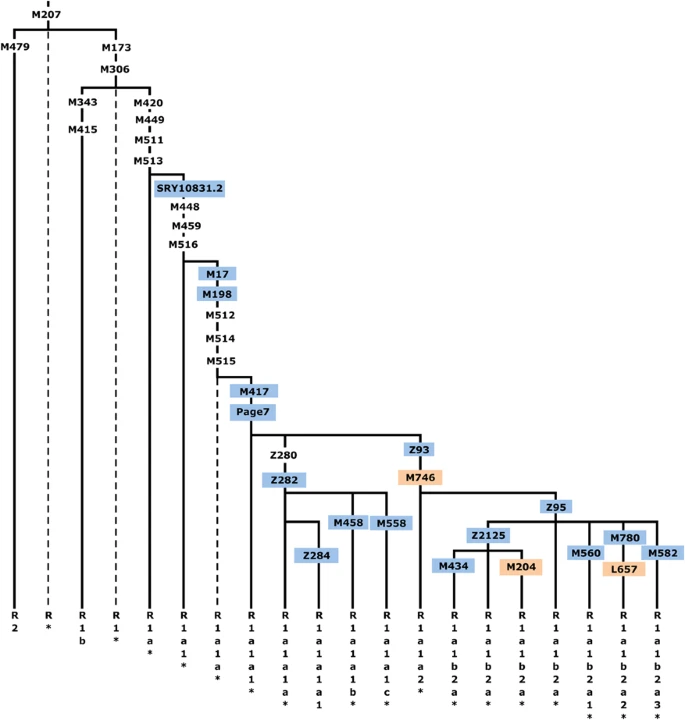 Figure 1 Haplogroup (hg) R1a-M420 topology, shown within the context of hg R-M207. Common names of the SNPs discussed in this study are shown along the branches, with those genotyped presented in color and those for which phylogenetic placement was previously unknown in orange. Hg labels are assigned according to YCC nomenclature principles with an asterisk (*) denoting a paragroup.63 Dashed lines indicate lineages not observed in our sample. The marker Z280 was not used as it maps to duplicated ampliconic tracts. More than 10% of men in a region extending from South Asia to Scandinavia share a common ancestor in hg R1a-M420, and the vast majority fall within the R1a1-M17/M198 subclade.22 Although the phylogeography of R1b-M343 has been described, especially in Western and Central Europe,15, 23, 24, 25 R1a1 has remained poorly characterized. Previous work has been limited to a European-specific subgroup defined by the single-nucleotide polymorphism (SNP) called M458.22, 26, 27, 28, 29, 30 However, with the discovery of the Z280 and Z93 substitutions within Phase 1 1000 Genomes Project data1 and subsequent genotyping of these SNPs in ∼200 samples, a schism between European and Asian R1a chromosomes has emerged.31 We have evaluated this division in a larger panel of populations, estimated the split time, and mapped the distributions of downstream sub-hgs within seven regions: Western/Northern Europe, Eastern Europe, Central/South Europe, the Near/Middle East, the Caucasus, South Asia, and Central Asia/Southern Siberia. Materials and methods Population samples We assembled a genotyping panel of 16 244 males from 126 Eurasian populations, some of which we report upon for the first time herein and others that we have combined from earlier studies,22, 29, 30, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 and updated to a higher level of phylogenetic resolution. All samples were obtained using locally approved informed consent and were de-identified. Whole Y-chromosome sequencing We analyzed 13 whole R1 Y-chromosome sequences: 8 novel, 2 previously published,4 and 3 from the 1000 Genomes Project.2 All sequences were generated on the Illumina HiSeq platform (Illumina, San Diego, CA, USA) using libraries prepared either from genomic DNA or from flow sorted Y-chromosomes drawn from lymphoblast cell line cultures induced to the metaphase cell division stage (Supplementary Table 1). We used Bowtie46 to map 101-bp sequencing reads to the hg19 human reference, and we called genotypes across 9.99 Mb and estimated coalescence times using a rate of 1 SNP accumulating per 122 years as described in Poznik et al.4 SNP analysis We selected binary markers (Supplementary Table 2) from the International Society of Genetic Genealogy10 database and from whole Y-chromosome sequencing and genotyped samples either by direct Sanger sequencing or RFLP assays. Within the full panel, 2923 individuals were found to be members of hg R1a-M420. These M420 carriers were then genotyped in a hierarchical manner (Figure 1) for the following downstream markers with known placement on the tree: SRY10831.2, M17, M198, M417, Page7, Z282/S198, Z284/S221, M458, M558/CTS3607, Z93/S202, Z95, Z2125, M434, M560, M780, M582, and for three SNPs whose placement within the R1a topology was previously unknown: M746, M204, and L657. We generated spatial frequency maps for the R1a subgroups that we determined to occur at 10% frequency or greater within a studied region. To do so, we used Surfer (v.8, Golden Software, Inc, Golden, CO, USA) with the kriging algorithm and the option to use bodies of water as break-lines. We carried out spatial autocorrelation analysis to detect clines by calculating Moran’s I coefficient using PASSAGE v.1.1 (www.passagesoftware.net) with a binary weight matrix, 10 distance classes, and the assumption of a random distribution. Haplogroup diversities were calculated using the method of Nei.47 To investigate the genetic affinities among populations, we used the freeware popSTR program (http://harpending.humanevo.utah.edu/popstr/) to perform a principal component analysis (PCA) based on R1a subgroup frequencies. Short tandem repeat (STR) analysis We genotyped 1355 samples for 10–19 STRs (Y-STRs; Supplementary Table 3) and calculated haplotype diversities, also using the method of Nei.47 Coalescence times (Td) of R1a subhaplogroups were estimated using the ASD0 methodology described by Zhivotovsky et al48 and modified according to Sengupta et al.41 Given the uncertainty associated with Y-STR mutation rates,24 we used both the evolutionary effective mutation rate of 6.9 × 10−4 per 25 years48 and, for comparison, the pedigree mutation rate of 2.5 × 10−3 per generation.49 |
|
|
|
Post by Admin on Dec 22, 2021 21:10:07 GMT
Results and Discussion Refinement of hg R1a topology Figure 1 shows, in context, the phylogenetic relationships of the markers we genotyped in this study. These include three for which phylogenetic placement was previously unknown: M746, M204, and L657. We localized the rare M204 SNP based on a single Iranian sample confirmed by Sanger sequencing to carry the derived allele.37, 50 Phylogeography We measured R1a haplogroup frequency by population (Supplementary Table 4). Of the 2923 hg R1a-M420 samples, 2893 were derived for the M417/Page7 mutations (1693 non-Roma Europeans and 1200 pan-Asians), whereas the more basal subgroups were rare. We observed just 24 R1a*-M420(xSRY10831.2), 6 R1a1*-SRY10831.2(xM198), and 12 R1a1a1-M417/Page7*(xZ282,Z93). We did not observe a single instance of R1a1a-M198*(xM417,Page7), but we cannot exclude the possibility of its existence. Of the 1693 European R1a-M417/Page7 samples, more than 96% were assigned to R1a-Z282 (Figure 2), whereas 98.4% of the 490 Central and South Asian R1a lineages belonged to hg R1a-Z93 (Figure 3), consistent with the previously proposed trend.31 Both of these haplogroups were found among Near/Middle East and Caucasus populations comprising 560 samples. Figure 2 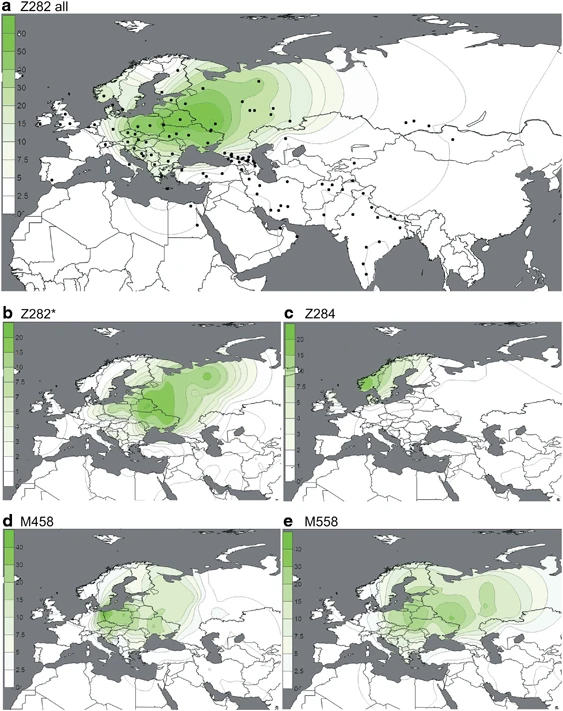 (a–e) Spatial frequency distributions of Z282 affiliated haplogroups. Each map was generated using the frequencies from Supplementary Table 4 among 14 461 individuals, distributed across 119 population samples (references listed in Supplementary Table 4). Because of the known difference between the origin and present distribution of the Roma and Jewish populations, they were excluded from the plots. Additional populations from literature27 were used for the M458 map. Figure 3 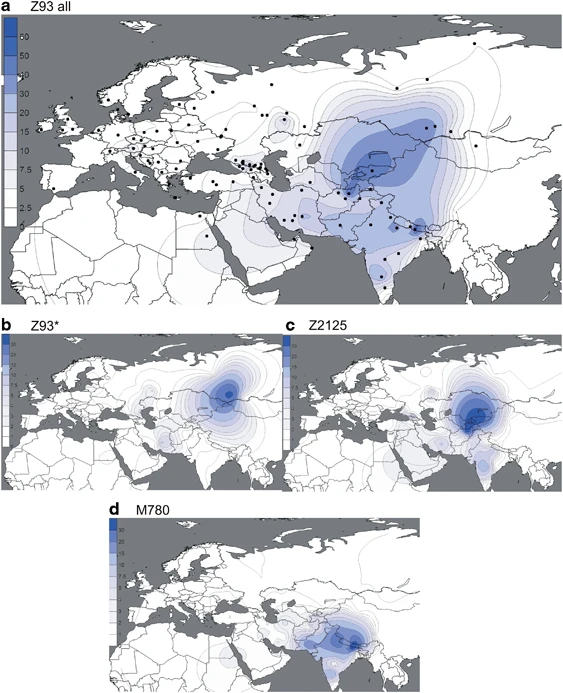 (a–d) Spatial frequency distributions of Z93 affiliated haplogroups. Maps were generated as described in Figure 2. Subgroups of both R1a-Z282 and R1a-Z93 exhibit geographic localization within the broad distribution zone of R1a-M417/Page7. Among R1a-Z282 subgroups (Figure 2), the highest frequencies (∼20%) of paragroup R1a-Z282* chromosomes occur in northern Ukraine, Belarus, and Russia (Figure 2b). The R1a-Z284 subgroup (Figure 2c) is confined to Northwest Europe and peaks at ∼20% in Norway, where the majority of R1a chromosomes (24/26) belong to this clade. We found R1a-Z284 to be extremely rare outside Scandinavia. R1a-M458 (Figure 2d) and R1a-M558 (Figure 2e) have similar distributions, with the highest frequencies observed in Central and Eastern Europe. R1a-M458 exceeds 20% in the Czech Republic, Slovakia, Poland, and Western Belarus. The lineage averages 11–15% across Russia and Ukraine and occurs at 7% or less elsewhere (Figure 2d). Unlike hg R1a-M458, the R1a-M558 clade is also common in the Volga-Uralic populations. R1a-M558 occurs at 10–33% in parts of Russia, exceeds 26% in Poland and Western Belarus, and varies between 10 and 23% in the Ukraine, whereas it drops 10-fold lower in Western Europe. In general, both R1a-M458 and R1a-M558 occur at low but informative frequencies in Balkan populations with known Slavonic heritage. The rarity of R1a-M458 and R1a-M558 among Central Asian and South Siberian R1a samples (4/301; Supplementary Table 4) suggests low levels of historic Slavic gene flow. In the complementary R1a-Z93 haplogroup, the paragroup R1a-Z93* (Figure 3b) is most common (>30%) in the South Siberian Altai region of Russia, but it also occurs in Kyrgyzstan (6%) and in all Iranian populations (1–8%). R1a-Z2125 (Figure 3c) occurs at highest frequencies in Kyrgyzstan and in Afghan Pashtuns (>40%). We also observed it at greater than 10% frequency in other Afghan ethnic groups and in some populations in the Caucasus and Iran. Notably, R1a-M780 (Figure 3d) occurs at high frequency in South Asia: India, Pakistan, Afghanistan, and the Himalayas. The group also occurs at >3% in some Iranian populations and is present at >30% in Roma from Croatia and Hungary, consistent with previous studies reporting the presence of R1a-Z93 in Roma.31, 51 Finally, the rare R1a-M560 was only observed in four samples: two Burushaski speakers from north Pakistan, one Hazara from Afghanistan, and one Iranian Azeri. |
|









