|
|
Post by Admin on Apr 13, 2022 18:35:02 GMT
The continent is the cradle of human origins and ingenuity, and with each new fossil and archaeological discovery, we learn more about our shared African past. Such research tends to focus on when our species, Homo sapiens, spread out to other landmasses 80,000–60,000 years ago. But what happened in Africa after that, and why don’t we know more about the people who remained? Our new study, conducted by an interdisciplinary team of 44 researchers based in 12 countries, helps answer these questions. By sequencing and analyzing ancient DNA (aDNA) from people who lived as long ago as 18,000 years, we roughly doubled the age of sequenced aDNA from sub-Saharan Africa. And this genetic information helps anthropologists like us understand more about how modern humans were moving and mingling in Africa long ago.  TRACING OUR HUMAN PAST IN AFRICA Beginning about 300,000 years ago, people in Africa who looked like us—the earliest anatomically modern humans—also started behaving in ways that seem very human. They made new kinds of stone tools and began transporting raw materials up to 250 miles, likely through trade networks. By 140,000–120,000 years ago, people made clothing from animal skins and began to decorate themselves with pierced marine shell beads. While early innovations appeared in a patchwork fashion, a more widespread shift happened around 50,000 years ago—around the same time that people started moving into places as distant as Australia. New types of stone and bone tools became common, and people began fashioning and exchanging ostrich eggshell beads. And while most rock art in Africa is undated and badly weathered, an increase in ochre pigment at archaeological sites hints at an explosion of art. TRACING OUR HUMAN PAST IN AFRICA Beginning about 300,000 years ago, people in Africa who looked like us—the earliest anatomically modern humans—also started behaving in ways that seem very human. They made new kinds of stone tools and began transporting raw materials up to 250 miles, likely through trade networks. By 140,000–120,000 years ago, people made clothing from animal skins and began to decorate themselves with pierced marine shell beads. While early innovations appeared in a patchwork fashion, a more widespread shift happened around 50,000 years ago—around the same time that people started moving into places as distant as Australia. New types of stone and bone tools became common, and people began fashioning and exchanging ostrich eggshell beads. And while most rock art in Africa is undated and badly weathered, an increase in ochre pigment at archaeological sites hints at an explosion of art. What caused this shift, known as the Later Stone Age transition, has been a longstanding archaeological mystery. Why would certain tools and behaviors, which up until that point had appeared in a piecemeal way across Africa, suddenly become widespread? Did it have something to do with changes in the number of people or how they interacted? THE CHALLENGE OF ACCESSING THE DEEP PAST Archaeologists reconstruct human behavior in the past mainly through things people left behind—remains of their meals, tools, ornaments, and sometimes even their bodies. These records may accumulate over thousands of years, creating views of daily livelihoods that are really averages over long periods of time. However, it’s hard to study ancient demography, or how populations changed, from the archaeological record alone. This is where DNA can help. When combined with evidence from archaeology, linguistics, and oral and written history, scientists can piece together how people moved and interacted based on which groups share genetic similarities. But DNA from living people can’t tell the whole story. African populations have been transformed over the past 5,000 years by the spread of herding and farming, the development of cities, ancient pandemics, and the ravages of colonialism and slavery. These processes caused some lineages to vanish and brought others together, forming new populations. Using present-day DNA to reconstruct ancient genetic landscapes is like reading a letter that was left out in the rain: Some words are there but blurred, and some are gone completely. Researchers need ancient DNA from archaeological human remains to explore human diversity in different places and times, and to understand what factors shaped it. Unfortunately, aDNA from Africa is particularly hard to recover because the continent straddles the equator and heat and humidity degrade DNA. While the oldest aDNA from Eurasia is roughly 400,000 years old, all sequences from sub-Saharan Africa to date have been younger than around 9,000 years. |
|
|
|
Post by Admin on Apr 13, 2022 21:16:42 GMT
Ancient DNA and deep population structure in sub-Saharan African foragers
Mark Lipson, Elizabeth A. Sawchuk, …Mary E. Prendergast
Nature volume 603, 290–296 (2022)
Abstract
Multiple lines of genetic and archaeological evidence suggest that there were major demographic changes in the terminal Late Pleistocene epoch and early Holocene epoch of sub-Saharan Africa1,2,3,4. Inferences about this period are challenging to make because demographic shifts in the past 5,000 years have obscured the structures of more ancient populations3,5. Here we present genome-wide ancient DNA data for six individuals from eastern and south-central Africa spanning the past approximately 18,000 years (doubling the time depth of sub-Saharan African ancient DNA), increase the data quality for 15 previously published ancient individuals and analyse these alongside data from 13 other published ancient individuals. The ancestry of the individuals in our study area can be modelled as a geographically structured mixture of three highly divergent source populations, probably reflecting Pleistocene interactions around 80–20 thousand years ago, including deeply diverged eastern and southern African lineages, plus a previously unappreciated ubiquitous distribution of ancestry that occurs in highest proportion today in central African rainforest hunter-gatherers. Once established, this structure remained highly stable, with limited long-range gene flow. These results provide a new line of genetic evidence in support of hypotheses that have emerged from archaeological analyses but remain contested, suggesting increasing regionalization at the end of the Pleistocene epoch.
Main
Models for the expression of human behavioural complexity during the Late Pleistocene (around 125–12 thousand years ago (ka)) often invoke demographic change1,2. By around 50 ka, technological innovations and symbolic behaviours (such as ornaments, bone tools, pigments and microliths) that were present earlier in the Middle Stone Age (MSA) become more consistently expressed across sub-Saharan Africa4,6,7. Archaeologists refer to this as the transition to the Later Stone Age (LSA)1,7,8,9. By around 20 ka, these material culture components were nearly ubiquitous, but regionally diverse. One explanation is that people began living in larger and/or more connected groups, with variations in population size and connectivity driving differences in material culture across space and time. Given the morphological variation among Late Pleistocene skeletons, interactions may have involved deeply structured populations2,10, consistent with some population history models based on genetics3.
The advent of genome-wide ancient DNA (aDNA) technology holds promise for better understanding major changes in material culture and hypothesized demographic shifts among ancient African foragers (Supplementary Notes 1, 2). Compared to elsewhere, especially Europe, there has been little genomic investigation of ancient African peoples. Previously available aDNA sequences from sub-Saharan African foraging contexts11,12,13,14, despite being relatively recent (younger than about 9 ka), provide evidence of ancient genetic structure that has since been disrupted by demographic transformations (such as the spread of food production, as well as colonialism, imperialism, enslavement and modern sociopolitical reorganization). The structure of ancient populations cannot be robustly reconstructed based solely on genetic data from present-day people.
Here we present new genome-wide aDNA data and radiocarbon dates from three Late Pleistocene and three early to middle Holocene individuals associated with LSA technologies at five sites in eastern and south-central Africa: Kisese II and Mlambalasi Rockshelters in Tanzania, Fingira and Hora 1 Rockshelters in Malawi, and Kalemba Rockshelter in Zambia (Fig. 1a and Extended Data Table 1). Direct and indirect dates range from around 18 ka to 5 ka, doubling the time depth of aDNA reported from sub-Saharan Africa. We analyse these data together with the published sequences of 28 other ancient African individuals recovered from contexts spanning the past 8,000 years and largely associated with foraging at 17 sites in eastern, central and southern Africa. We also provide higher-coverage data for 15 of these individuals. Analysis of the ancient data together with sequences from present-day groups, aided by new statistical methods, enables a reconstruction of changes in regional- and continental-scale population structures among people who lived before the sweeping demographic changes of the past approximately 5,000 years. It also enables comparisons of Pleistocene forager population dynamics between the tropics and more temperate regions.
|
|
|
|
Post by Admin on Apr 14, 2022 1:14:01 GMT
Fig. 1: Locations of the individuals analysed and PCA analysis. 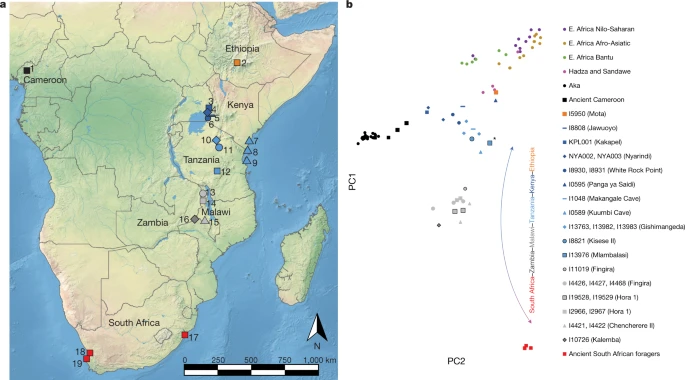 a, Locations of individuals analysed in this study. The shapes and colours of the symbols correspond to the PCA in b. 1, Shum Laka; 2, Mota Cave; 3, Kakapel RS (Rockshelter); 4, Nyarindi RS; 5, Jawuoyo RS; 6, White Rock Point; 7, Panga ya Saidi; 8, Makangale Cave; 9, Kuumbi Cave; 10, Gishimangeda Cave; 11, Kisese II RS; 12, Mlambalasi RS; 13, Fingira; 14, Hora 1; 15, Chencherere II; 16, Kalemba RS; 17, Ballito Bay; 18, Faraoskop RS; 19, St Helena. b, PCA results. Axes were computed using present-day groups from eastern (Dinka pastoralists), southern (Juǀ'hoansi foragers) and central Africa (Mbuti foragers). Small circles represent present-day individuals; other symbols represent ancient individuals (larger points corresponding to earlier individuals and black outlines to newly reported individuals). The lowest-coverage individual (from Mlambalasi), shown with an asterisk, has the most uncertain position. The base map in a is from Natural Earth (https://www.naturalearthdata.com). E., east. The dataset Of 31 samples (Supplementary Table 1), five petrous bones and one distal phalanx yielded aDNA sequences, which, after preparation of up to six libraries from each sample and enrichment for a panel of around 1.2 million single-nucleotide polymorphisms (SNPs), ranged in coverage from 0.001–3.2× (median, 0.06×) of targeted genome-wide SNP positions (Extended Data Table 1 and Supplementary Table 2). Additional archaeological and bioarchaeological information for these individuals is summarized in Supplementary Note 3. Direct 14C dates were attempted for the five petrous bones, but only two preserved sufficient collagen: Kalemba (I10726; 5,280–4,880 calibrated years before present (cal. BP), PSUAMS-4764) and Kisese II (I18821; 7,240–6,985 cal. BP, PSUAMS-4718) (Supplementary Table 3 and Supplementary Note 4). Moreover, a new date was generated on enamel carbonate for a published individual from Hora 1 (I2966; previously estimated around 8,100 BP, now directly dated to 9,090–8,770 cal. BP, PSUAMS-5145). Individuals from Mlambalasi (I13976; about 20–17 ka) and Hora 1 (I19528, I19529; 17–14 ka) are well constrained to the Late Pleistocene based on multiple indirect dates (Supplementary Table 4 and Supplementary Note 3). One individual from Fingira (I11019) is represented by a distal phalanx that was recovered in isolation near the surface during excavation. This sample was too small to be both dated and assessed for aDNA; its age is constrained to around 6,200–2,300 cal. BP by association with direct dates on other human remains from the site. The 15 previously published individuals11,13,15,16 (Supplementary Note 3) for which we increase sequence coverage include approximately 26× shotgun coverage for the individual from Mota Cave in Ethiopia15 (I5950), enabling reliable calling of diploid genotypes (Extended Data Table 1, Methods and Supplementary Table 2). The authenticity of the new aDNA data was assessed through a combination of several criteria; detectable contamination was observed for only two samples (Methods, Extended Data Fig. 1a, Supplementary Table 2 and Supplementary Note 5). In Supplementary Table 5 and Supplementary Note 5, we report genotypes at SNPs associated with lactase persistence, sickle cell trait and the Duffy antigen, with derived alleles observed only at the DARC (Duffy) locus (four published individuals from Cameroon). Uniparental markers All four newly reported males are similar to most published ancient foragers from this region of Africa in carrying the widely distributed Y chromosome haplogroup B2 (Extended Data Table 1). Among the 23 individuals in our dataset with known mtDNA haplogroups, up to 14—almost all from Kenya and Tanzania—have haplogroups that are today associated with eastern Africa (Extended Data Table 1 and Supplementary Table 6). Eight individuals—all from Malawi and Zambia—have haplogroups that are associated with some ancient and present-day southern African people, specifically groups for whom foraging is the main mode of subsistence17,18,19,20. Two individuals from Malawi (I19529 from Hora 1, dating to about 16 ka and carrying L5b, and I4426 from Fingira, dating to about 2.3 ka and carrying L0f/L0f3) have eastern-Africa-associated haplogroups, whereas a different individual from Malawi (I2967 from Hora 1, dating to about 8.2 ka with L0a2/L0a2b) and possibly one from Kenya (I8930 from White Rock Point with L2a4) belong to lineages that are characteristic of present-day central African foragers (such as Mbuti and Aka). These results show that eastern and south-central Africa was home to, and an area of interaction among, diverse ancient foraging groups, and also that several of these haplogroup lineages were formerly more widespread than they are today. |
|
|
|
Post by Admin on Apr 14, 2022 17:17:38 GMT
Three-way cline of genome-wide ancestry
For the bulk of our analyses, we used the genome-wide genotype data to gain insights into the ancestry of the ancient forager individuals and their connections to other groups. We performed a supervised principal component analysis (PCA) (Methods) in which we used three present-day groups—Juǀ'hoansi (San) from southern Africa, Mbuti from central Africa and Dinka from northeastern Africa—to define a two-dimensional plane of variation, and projected all other individuals (ancient and present day) onto this plane (Fig. 1b). Consistent with previous studies5,11,13,14, we observed an ancient latitudinal gradient of ancestry, represented at its northern extreme by an individual from around 4.5 ka from Mota Cave,
Ethiopia, and its southern extreme by individuals from around 2 ka from South Africa. The newly reported individuals generally cluster with their geographical neighbours but extend documentation of the cline both geographically (southwest to Kalemba, at the corresponding extreme on PCA) and temporally (to a maximum of approximately 18–16 ka, with no apparent temporal subclusters). Furthermore, we found complexity in the cline in the form of deviations from a straight line: (1) the main direction of variation does not align with ancient southern African foragers; and (2) several individuals appear to shift in the direction of present-day and ancient central African foragers. Both observations may indicate that some of the ancient eastern and south-central African individuals sampled here trace part of their ancestry to groups that are related to foragers currently living in central Africa. Furthermore, (1) could indicate that the southern-African-related ancestry among the ancient individuals is only distantly related to present-day Juǀ'hoansi and ancient southern African foragers.
We used allele-sharing tests (f-statistics) (Methods) to further investigate which individuals differed in their degree of relatedness to ancient South African foragers (AncSA) (Extended Data Table 1), the Mota individual or present-day Mbuti. Consistent with the PCA, most pairs of individuals from the same region (including from different time points) were nearly symmetric in their ancestry (|Z| < 3) (Supplementary Table 7). The exceptions were (1) excess affinity between Mbuti and KPL001 (Kakapel; max Z = 5.1); (2) excess affinity between AncSA and I0589 (Kuumbi Cave; max Z = 4.1); and (3) modest differences within Malawi and Zambia (max Z = 3.8). By contrast, well-powered cross-region statistics were highly significantly non-zero, for example, f4(I8808 (Jawuoyo), I8821 (Kisese II); Mota, AncSA) > 0, Z = 7.8. We also used the qpWave program in ADMIXTOOLS to combine multiple f-statistic-based signals into a test for the number of distinct components of ancestry (relative to a specified outgroup set) present among the (sampled) ancient forager individuals (Methods). We found that at least three sources are
necessary (P = 6.4 × 10−14 for rejecting a two-source model) but, interestingly (with our available statistical power), that three sources are also sufficient (P = 0.73; four versus three sources P = 0.15), even with Mota, San (here, both Juǀ'hoansi and ǂKhomani) and Mbuti among the outgroups. When we added the Mota individual to the test set, we found increased evidence for a fourth source, despite the less stringent outgroups (P = 0.07; four versus three sources P = 0.019) (Methods). This result could reflect a highly divergent ancestry component contributing to the Mota individual inferred in previous work16; additional lineages may also have been present among as-yet unsampled ancient individuals from these regions.
We attempted to estimate the dates of admixture (potentially involving any distinct sources of ancestry) for the ancient foragers using DATES21. With the caveat that our power is limited by data availability, we obtained only two robust estimates (Supplementary Table 8), both for previously published individuals, and both (given the additional results below) are probably connected to admixture from food producers: for I4421 (Chencherere II, no direct age, past approximately 5,000 years), a date of 10 ± 2 generations before the individual lived; and for I1048 (Makangale Cave, direct age, past approximately 1,500 years), 79 ± 24 generations before the individual lived.
|
|
|
|
Post by Admin on Apr 14, 2022 19:26:16 GMT
Inter- and intraregional relationships Next, we modelled the ancestry of the ancient foragers in an admixture graph framework to test additional hypotheses concerning their relationships, aided by a new methodology to increase available information from low-coverage data (Figs. 2 and 3, Methods, Supplementary Notes 6 and 7 and Extended Data Figs. 2–5). In model 1, along with other populations, we included three geographically and genetically diverse ancient eastern and south-central African individuals with high sequencing coverage: I4426 (Fingira, about 2.5 ka), I8821 (Kisese II) and I8808 (Jawuoyo). On the basis of the results in the previous section, we hypothesized that they could be fit with mixtures of three ancestry components: one related to the Mota individual (representing an ancient group of foragers from the northern part of eastern Africa), one related to central African foragers (represented by present-day Mbuti) and one related to southern African foragers (represented by four ancient individuals from South Africa). Indeed, we obtained a good fit to the data in model 1 (max residual Z = 2.0), even when specifying identical sources for all three individuals, and the relative ancestry proportions were as expected: Mota-related ancestry decreased from north to south, and Jawuoyo (I8808) had the highest ratio of central-African-related ancestry to southern-African-related ancestry. Omitting any of the three components for any of the individuals results in a poor fit (Z ≥ 4.0) (Supplementary Note 6). As in ref. 16, we also estimated around 30% of a separate and deeply diverged ‘ghost’ ancestry component in the Mota individual (replicated here using new higher-coverage diploid whole-genome data). Fig. 2: Schematic of admixture graph results.  Branch lengths are not to scale. The arrows denote admixture events, with the three primary components of ancestry shown as dashed arrows, and other inferred gene flow as small solid arrows (with colours corresponding to related groups). Subclusters of ancient eastern and south-central African foragers reflect the inferred instances of excess relatedness among individuals, with internal branch lengths shown in genetic-drift units. Mixture proportions are shown in Fig. 3 and Supplementary Table 9 and the full results are shown in Extended Data Fig. 4. Individual laboratory numbers are shown at the bottom (Extended Data Table 1). N., north; W., west. Fig. 3: Distribution of main ancestry components. 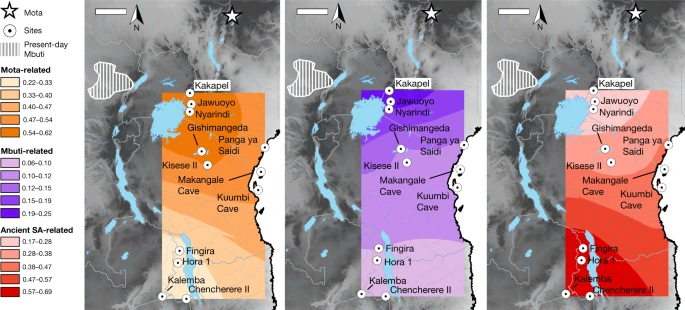 Kriged distribution of the proportions of each of the three main ancestry components (summing to 1) found in ancient eastern and south-central African foragers analysed in this study (details are provided in Supplementary Table 9). The approximate present-day Mbuti home region is from ref. 41. Individuals from the same site were included using locations that differed by 0.000001 decimal degrees latitude to ensure representation in the interpolation. Scale bars, 250 km. Topographical data are from the Shuttle Radar Topography Mission (SRTM)42. SA, southern African. |
|





