|
|
Post by Admin on Feb 16, 2021 20:32:18 GMT
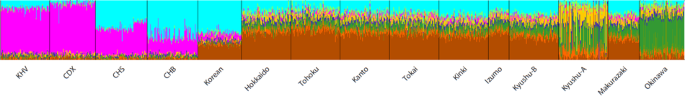 Fig. 4 Admixture analysis for k (number of ancestral components) = 9. For clarity, only 100 randomly selected individuals from each region of the BioBank Japan dataset are included in this figure An unrooted tree was constructed using the neighbor-joining method [25] based on Fst distances between populations (Supplementary Fig. 4). Populations are located more or less in a linear fashion; Okinawa/Kyushu-A cluster on the left and the CHB/Korean cluster at the right. Yamato people are in the center, with Kinki being the closest to the CHB/Korean cluster. We also constructed phylogenetic networks using Neighbor-Net method [26] focusing on Yaponesians and Korean/CHB (Fig. 5) and their relationship with other continental Asians (Supplementary Fig. 5). Because the Ainu population is very distant from the remaining populations (Fig. 5A), they were omitted from the subsequent network (Fig. 5B). Now Okinawa and Kyushu-A populations formed one cluster, and the remaining seven Yaponesian populations (shown in light blue background in Fig. 5B) are located between this cluster and the Korean-CHB cluster. Figure 5c shows the phylogenetic relationship of these seven Yaponesian populations. It should be noted that BBJ data for Fig. 5 were sampled in a different way than that for the remaining figures; 100 individuals were randomly chosen from seven populations of BioBank. This sampling difference probably caused the change of the Tohoku population; either somewhat closer to Izumo in Fig. 5C or almoThe results of f4 test for gene flow are listed in Table 1. To test if there is gene flow between Yaponesians and continental Asians, we tested f4(GBR, CHB; Okinawa, x) where x is the test population. Consistent with PCA, phylogenetic trees and network, Kinki showed the highest Z-score (26.2) among the Yamato populations. Similarly, f4(GBR,CHB;x1,x2) where x1 and x2 are non-Okinawan Yaponesians also showed evidence of gene flow between Kinki and CHB (Supplementary Table). For example, most significant Z-scores were observed for f4(GBR,CHB; Kanto, Kinki), Z-score: 24.7. We also tested f4(GBR, x; Izumo, Kinki) to test if there is gene flow involving Izumo. Negative f4 values indicative of gene flow was observed for Okinawa, Kyushu-A, and Makurazaki, although the Z-scores are relatively low. This was repeated with Makurazaki instead of Izumo using f4(GBR, x; Kinki, Makurazaki), and results suggest presence of gene flow between Makurazaki and Okinawa or Kyushu-A. Gene flow between Okinawa and Kyushu-A was further supported by f4(GBR, Okinawa; x1, x2) where x1 and x2 are non-Okinawa Yaponesians (Supplmenentary Table).
Table 1 f4 test for gene flow
From: Genome-wide SNP data of Izumo and Makurazaki populations support inner-dual structure model for origin of Yamato people
x f4 Std. error Z-score
f4(GBR, CHB; Okinawa, x)
Hokkaido 0.00124 5.07E−05 24.49
Tohoku 0.00111 4.79E−05 23.16
Kanto 0.00124 5.06E−05 24.48
Tokai 0.00127 5.17E−05 24.58
Kinki 0.00143 5.44E−05 26.20*
Izumo 0.00100 7.12E−05 14.08
Kyushu-A 0.00003 2.16E−05 1.16
Kyushu-B 0.00124 4.94E−05 25.12
Makurazaki 0.00111 5.82E−05 19.05
f4(GBR, x; Izumo, Kinki)
Hokkaido 0.00008 4.89E−05 1.58
Tohoku 0.00005 4.92E−05 0.97
Kanto 0.00007 4.90E−05 1.44
Tokai 0.00009 4.89E−05 1.74
Kyushu-A −0.00008 5.14E−05 −1.63
Kyushu-B 0.00009 4.89E−05 1.78
Makurazaki −0.00003 5.00E−05 −0.69
Okinawa −0.00008 5.12E−05 −1.64*
f4(GBR, x; Kinki, Makurazaki)
Hokkaido −0.00015 4.28E−05 −3.49
Tohoku −0.00013 4.29E−05 −3.03
Kanto −0.00014 4.28E−05 −3.33
Tokai −0.00015 4.28E−05 −3.46
Izumo −0.00002 4.50E−05 −0.41
Kyushu-A 0.00020 4.44E−05 4.45
Kyushu-B −0.00010 4.27E−05 −2.35
Okinawa 0.00022 4.38E−05 4.92*
st identical to JPT in Supplementary Fig. 5. Makurazaki is the closest to the Okinawa and Kyushu-A cluster (indicated by splits labeled b, c, and d), followed by Izumo as shown by split a. Okinawa, Kyushu-A, and Tohoku are separated from the remaining nine populations by split e. Fig. 5 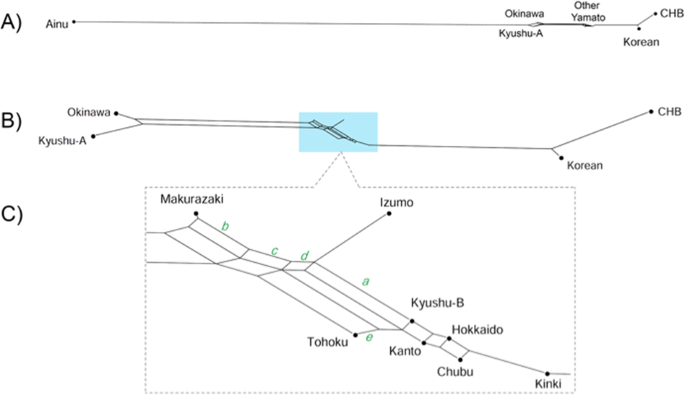 |
|
|
|
Post by Admin on Feb 16, 2021 21:27:10 GMT
Discussion We generated genome-wide SNP data for Izumo in the San-in region and Makurazaki in the southern Kyushu region (see Fig. 1) using Japonica array. The genetic location of Makurazaki population in the PCA plot (Fig. 3) and phylogenetic network (Fig. 5) is more or less similar to the geographical location of this city, namely, between Okinawa and the mainland. This is supported by f4(GBR, x; Kinki, Makurazaki) test showing Makurazaki having more gene flow with Okinawa and Kyushu-A than with other Yamato. Geographically, Izumo city is facing the sea between Yaponesia and Continental East Asia (Fig. 1). The distance to the Korean Peninsula is ~350 km, and it was expected that Izumo people may be closer to continental Asians. However, f4 test showed that the Kinki population is the closest to continental Asians (f4(GBR, CHB; Okinawa, x)). This is supported by another study using genome-wide SNPs [29]. Interestingly, Izumo population has higher genetic affinity to Okinawa and Kyushu than to other populations in Honshu, as suggested by f4(GBR, x; Izumo, Kinki). This discrepancy suggests an existence of some gene flow events that are different from usually expected isolation by distance [30]. Yamaguchi-Kabata et al. [9] analyzed genome-wide SNP data of 7,000 Japanese collected from seven geographical regions (Hokkaido, Tohoku, Kanto, Tokai, Kinki, Kyushu, and Okinawa) as part of the BBJ Project. They found the two clear clusters called Hondo and Ryukyu, which we also observed (Fig. 3). We also confirmed region specific characteristics discovered by Yamaguchi-Kabata et al. [9] and later confirmed by Okada et al. [31] in which Hokkaido, Kanto, and Tokai area individuals are more or less genetically similar, whereas Tohoku individuals are slightly differentiated from those three regions people based on PCA, network and f4 tests. We also confirm that Kinki people are the closest to continental Asians as reported by Yamaguchi-Kabata et al. [9] and Watanabe et al. [29]. Kinki area includes Nara and Kyoto, past capitals of Japan and thus has a history of human migration from the Asian continent. These results and HLA data [11] prompted Saitou [32] to propose the three-migration model. Later, Saitou and Jinam [13] and Saitou [2] proposed the “Inner-Dual Structure” model of Yaponesians, shown in Fig. 6. After the initial peopling of the Japanese Archipelago by the ancestors of Jomon and possibly including the paleolithic period people (first migration), there were possibly two (or more) human migrations from the Asian continent. Although the timing of this event is not yet determined, these people may have occupied coastal areas of the main island Honshu (“Periphery” in Fig. 6). Then the third migration from the continent mostly involved movement between the Asian Continent and six areas (Hakata in northern Kyushu, Osaka, Kyoto, Nara in Kinki, Kamakura, and Edo-Tokyo in Kanto; shown in gray circles in Fig. 6). These “Central Axis” areas were historically the cultural and political centers in Yaponesia, and are assumed to attract many new migrants from continental East Asia. 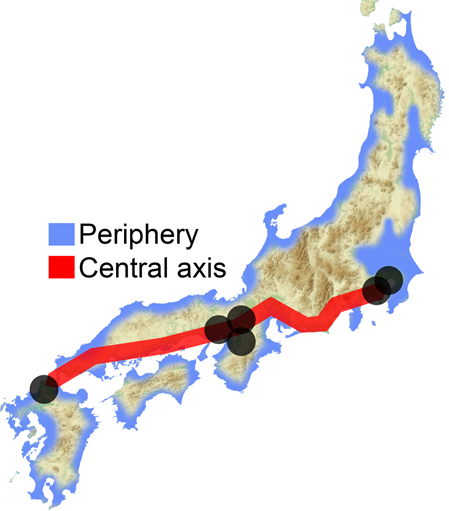 Fig. 6 Two additional data also supported the “Inner-Dual Structure” model according to Saitou [2]; five mtDNA haplogroup frequency data of 18,641 Yaponesians sampled from all the 47 prefectures provided by Genesis Healthcare (see Supplementary Fig. 6; taken from Fig 52 of Saitou [2]) and ABO blood group allele frequency data of 46 prefectures except for Okinawa by Fujita et al. [33] and those for Okinawa Prefecture [34, 35] (see Supplementary Fig. 7; taken from Figure 53 of Saitou [2]). We divided the 47 prefectures into 26 peripheral (shown in blue) and 21 central axis (shown in red) in both data. In Supplementary Fig. 6, Principal Component 1 broadly separates peripheral (right side) and central axis (left side) populations. This bias is statistically significant at the 1% level using Fisher’s exact test (see the upper right 2 × 2 table of Supplementary Fig. 6). The frequency plot of alleles A and B of the ABO blood group (Supplementary Fig. 7) tend to group peripheral prefectures (shown in blue) to the lower left of the diagonal dotted line, and this bias is also statistically significant at the 1% level using Fisher’s exact test (see the upper right 2 × 2 table of Supplementary Fig. 7). Recently, HLA data for all 47 prefectures were analyzed [12], and a phylogenetic relationship of 15 regions was shown. Interestingly, populations in northern Kyushu and southern Kanto are phylogenetically closer to continental Asian populations, whereas northern Tohoku, Shikoku and Okinawa formed one cluster. These patterns are compatible with the “Inner-Dual Structure” model shown in Fig. 6. However, San-in (belonging to Periphery in Fig. 6) and San-yo (belonging to Central Axis in Fig. 6) populations in Chugoku region are clustered in the phylogenetic tree of Hashimito et al. [12]. The “Inner-Dual Structure” model might explain why people in the Central Axis region appear closely related in the PCA and network analyzes, whereas people from the Periphery such as Makurazaki, Izumo, or Tohoku were less genetically influenced by the more recent migrations. However it is still not clear when those new and old migrants came to Yaponesia. Overall, these results appear to support the “Inner-Dual Structure” model, however other explanations such as very recent migrations might contribute to the observed patterns. The addition of samples from under-represented regions in Japan together with whole genome sequence data is needed to properly decipher the genetic make-up of Yaponesians. |
|
|
|
Post by Admin on Aug 8, 2021 22:26:02 GMT
Japan considered from the hypothesis of farmer/language spread Published online by Cambridge University Press: 05 May 2020 Formally, the Farming/Language Dispersal hypothesis as applied to Japan relates to the introduction of agriculture and spread of the Japanese language (between ca. 500 BC–AD 800). We review current data from genetics, archaeology, and linguistics in relation to this hypothesis. However, evidence bases for these disciplines are drawn from different periods. Genetic data have primarily been sampled from present-day Japanese and prehistoric Jōmon peoples (14,000–300 BC), preceding the introduction of rice agriculture. The best archaeological evidence for agriculture comes from western Japan during the Yayoi period (ca. 900 BC–AD 250), but little is known about northeastern Japan, which is a focal point here. And despite considerable hypothesizing about prehistoric language, the spread of historic languages/ dialects through the islands is more accessible but difficult to relate to prehistory. Though the lack of Yayoi skeletal material available for DNA analysis greatly inhibits direct study of how the pre-agricultural Jōmon peoples interacted with rice agriculturalists, our review of Jōmon genetics sets the stage for further research into their relationships. Modern linguistic research plays an unexpected role in bringing Izumo (Shimane Prefecture) and the Japan Sea coast into consideration in the populating of northeastern Honshu by agriculturalists beyond the Kantō region. Introduction The Jōmon period (see Figure 1) is one of the longest post-Palaeolithic archaeological periods in the world, sustained by a hunting–gathering–fishing–horticultural subsistence pattern (Habu, 2004). The introduction of rice agriculture into the Genkai Bay area of North Kyushu (Figure 1) from around 1000 BC marks the beginning of the Yayoi period. Wet-rice technology was introduced by Mumun (‘plain pottery’) culture migrants from the southern Korean Peninsula relatively early, ca. 1000 BC, in the Peninsula's Mumun period (1500–300 BC). These migrants were not Yayoi, they were Mumun. 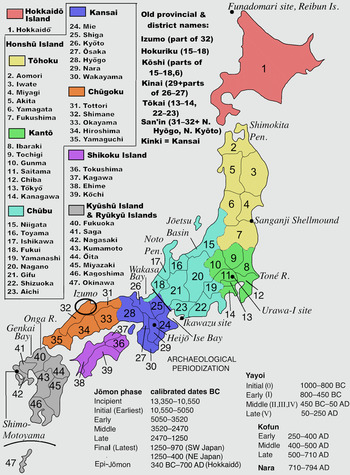 Figure 1. Map of Japan: prefectures, districts, and placenames mentioned in the text, with Japanese archaeological periodizations. Sources: Periodization from Barnes (2015), tables 1.6, 1.8. Map by TheOtherJesse [public domain], via Wikimedia Commons, modified by GLB [https://commons.wikimedia.org/wiki/File:Regions_and_Prefectures_of_Japan.svg] The term Yayoi has four uses, which can create much confusion. First, it is the designation of the period beginning with the introduction of rice agriculture around 1000 BC until the advent of the Mounded Tomb Culture in the third century AD. Yayoi is a period designation exclusive to Japan; it includes both farmers and hunter–gatherers and entails the agricultural transition in a time-transgressive and regionally disparate process. Second, ‘Yayoi people’ may refer to anyone living in the Japanese Islands in the Yayoi period, or third, Yayoi may refer specifically to admixed people (Mumun + Jōmon in varying in proportions and across great distances). Fourth, Yayoi may indicate acculturation: the adoption of (rice) agriculture (and other continental material culture) by Jōmon-lineage people in the Yayoi period. All of these conflicting aspects of Yayoi must be kept in mind and clearly defined in any discussion. Upon the Mumun migrants’ arrival, it is assumed from artefactual, skeletal morphological, and genetic evidence that many took partners from the resident Jōmon population, leading to mixed descendants – and to cultural transformation. These generational ‘admixed’ farmers then spread out from North Kyushu into the rest of the Japanese Islands, meeting more local Jōmon-lineage peoples with further admixing taking place. It is common in the literature to label the admixed farmers as ‘Yayoi’ because they carried the new culture based on rice, while increasingly recognizing that Jōmon-lineage peoples (distinguished as Epi-Jōmon in the north, but included within ‘Yayoi’ in the south) survived well into historical times without substantial admixture. The mixing of these two populations is recognized as the Dual Structure Hypothesis of the modern mainland Japanese population (Hanihara, 1991, 2000). The first section below reports on Jōmon genetics as a basis for further research on Yayoi admixtures. Despite the 13,000-year Jōmon history entailing multiple cultural formations and temporal trends, archaeologists tend to treat the Jōmon as a homogeneous people unadmixed with mainland East Asians and Austronesians since their separation from each other in the Palaeolithic. Genetic research is disabusing us of that approach, revealing potential Jōmon contacts with coastal mainland East Asians before the arrival of Mumun migrants; these data mirror archaeological evidence of contact. This is enlightening for its own sake, particularly in tracing connections between the Jōmon and continental peoples. However, in terms of investigating Yayoi admixtures, it makes the issue much more complicated. As more Yayoi-period genomic data becomes available, we may then be able to track the regional spread of continental genes through the Japanese archipelago. The addition of new Yayoi-period genomic data is vital, as new waves of immigration from the Korean Peninsula in the succeeding Kofun and Nara periods have enhanced the continental contribution in the modern Japanese, making it difficult to use present-day genomes to effectively study population movement and admixture in the Yayoi period. The second section focusses on the spread of agriculture from west to east through the Japanese Islands. The major topic of study in this spread has been the adoption of irrigated wet-rice agriculture. It has been assumed that this technology was spread by admixed farmer-migrants. In contrast, new studies are documenting instances of non-irrigated (swamp, dryland) rice and millet cultivation that may be earlier (e.g. Nasu & Momohara, 2016; Shitara & Takashe [sic], 2014). These authors assume that local Final Jōmon and Epi-Jōmon peoples were the cultivators, although some may have become farmers of irrigated wet rice later. The relationships between cultivation technologies and ethnic populations are often unclear, making the next step in the analysis, on language spread, difficult. The third section deals with language spread. Prehistoric language as such is inaccessible, but the fact that nowadays all languages on the islands except Ainu are related to Japanese makes it absolutely clear that the population that brought wet-rice agriculture to Japan must have been speakers of proto-Japanese. (Proto-Japanese is the ancestral language from which all modern varieties of Japanese descended.) Proto-Japanese split into several dialects, and then Ryūkyūan split off from the Kyūshū dialect. Evidence for the latter scenario has been growing in recent years. (For an overview see De Boer 2017.) Although the Ryūkyūan varieties form independent languages now, their split from the Kyūshū dialect makes them in origin dialects of Japanese. Proto-Japanese is postulated to have developed on the continent (Whitman, 2011; Miyamoto, 2016; Robbeets, 2005, 2017), and it is hypothesized to have had spread throughout the archipelago with the admixed Yayoi people practising irrigated rice agriculture. This accords well with the farming/language dispersal hypothesis (Renfrew, 1987; Bellwood & Renfrew, 2002). Problems, however, include exactly when and how this spread took place, whether it was indeed only associated with rice agriculture or also with other grain crops, and what Jōmon or Epi-Jōmon language(s) might have been replaced or pushed out by which dialect of Japanese. With the general lack of Yayoi DNA and written linguistic data, it is difficult to match food production techniques, genomic identity and language type. |
|
|
|
Post by Admin on Aug 9, 2021 2:39:48 GMT
Jōmon genetics
The transition from the Jōmon to the Yayoi periods marked a transition in the material culture of Japan, with the appearance and increase of tools, artefacts and archaeobotanical remains associated with agriculture. Studies have long associated this transition with the migration of Mumun people through the Korean peninsula into the closest island in the Japanese archipelago, Kyushu (Hammer & Horai, 1995; Hanihara, 1991). Early assessments using cranial and dental morphology and genetics of humans and their commensals led to the Dual Structure model (Hanihara, 1991), for which a key component is the argument that present-day Japanese are a mixture of populations indigenous to Japan since the Palaeolithic and later migrants from mainland Asia. Many studies, mostly using present-day genomes of indigenous populations in the archipelago (i.e. Ainu, Ryūkyū) as a proxy for Jōmon-related ancestry, found genetic patterns that suggest that the present-day Japanese populations are admixed, supporting that aspect of the Dual Structure model (Jeong, Nakagome, & Di Rienzo, 2016a; Jinam et al., 2015; Nakagome et al., 2015).
Advances in DNA sequencing methods have allowed access to genetic material for studying population demographic history in the Japanese archipelago. Uniparental markers were some of the earliest used to infer population history; these include mitochondrial DNA (mtDNA), which exists outside the nucleus, and the non-recombining portion of the Y-chromosome. The primary units considered when studying these markers are haplogroups, which are variants of a DNA segment that derives from the same common ancestor, and they can provide information on a population's genetic history.
Since uniparental transfer results in no recombination, it is straightforward to reconstruct phylogenetic trees and assign a haplogroup to the mtDNA or Y-chromosome region (Haber, Mezzavilla, Xue, & Tyler-Smith, 2016). Sets of haplogroups are now well annotated and easily identifiable in newly sequenced individuals using many tools and databases (e.g. ISOGG, 2019; Weissensteiner et al., 2016). If they are identified to be at high frequency in a localized area but in low frequency elsewhere, the origin of the haplogroup, or the common ancestor, can be inferred to have derived from that region. While the lack of recombination allows simple haplogroup assignation, it also means that it is a single data point representing population history, and stochastic events or sex-biased migration patterns can lead to inaccurate inference of the true population history (Nordborg, 1998; Rosenberg & Nordborg, 2002).
In contrast, the nuclear genome undergoes recombination, which means that small segments of each chromosome have effectively independent histories. Study of a single diploid genome (i.e. the 23 pairs of chromosomes in the cell nucleus) can be interpreted as study of small portions of thousands of different individuals, resulting in high statistical power for inferring the history of the population(s) represented by those individuals. Thus, sampling genome-wide is imperative for assessing the accuracy of the demographic relationships inferred from uniparental markers.
The ability to extract and sequence ancient DNA (aDNA) has revolutionized the study of human prehistory, allowing direct sampling of past populations and revealing genetic variation no longer observed today. Post-mortem degradation of DNA makes it very difficult to sequence aDNA, so the greater numbers of mtDNA makes it much easier to extract and sequence. The earliest ancient DNA studies in humans were for mtDNA (e.g. Krings et al., 1997), including from a Jōmon individual as early as 1989 (Horai et al., 1989) and Yayoi individuals as early as 1995 (Oota, Saitou, Matsushita, & Ueda, 1995). In fact, the majority of ancient DNA studies on Jōmon individuals have been on mtDNA (Schmidt & Seguchi, 2014).
Only recently have ancient nuclear genomes been retrieved from individuals from Japan. Ancient genomes have been published from human remains at different sites (Sanganji, Ikawazu, and Funadomari associated with Jōmon material culture; Shimo-Motoyama associated with Yayoi material culture; Figure 1). Two partial genomes sequenced from the Sanganji shellmound in Tōhoku were of 3000-year-old individuals (Kanzawa-Kiriyama et al., 2017). Genome-wide data have also been retrieved for a 2700-year-old individual from Ikawazu (McColl et al., 2018) and two 3900-year-old individuals from the Funadomori site in Hokkaido (Kanzawa-Kiriyama et al., 2019). Sampling of nuclear genomes is still sparse, but a rough understanding of the ancestry associated with the Jōmon is taking shape with each study.
Characterizing Jōmon-period ancestry is fundamental to understanding the genetic composition of humans that lived during the Yayoi period, who ostensibly represent various degrees of mixture between Jōmon and continental peoples. This in turn will improve our understanding of population migration on the archipelago and its role in the spread of agriculture across the archipelago. Here, we highlight some key features and emerging questions observed to date on Jōmon-related ancestry, which allow us to better understand the challenges (and rewards!) in interpreting the genetics of the Yayoi in future studies.
Analyses of both uniparental (Y and mtDNA) and nuclear DNA of present-day populations have highlighted that ancestry persists in Japan that is deeply diverged from ancestry found in mainland East Asian populations. The distribution of haplogroup frequencies across multiple populations is often used to determine where haplogroups originated. For example, those that occur in high frequency in present-day populations from Japan but low frequency elsewhere (e.g. N9b, M7a, G1b) were proposed as candidate haplogroups of Jōmon origin (Adachi, Shinoda, Umetsu, & Matsumura, 2009; Adachi et al., 2011; Kanzawa-Kiriyama, Saso, Suwa, & Saitou, 2013; Kivisild et al., 2002).
The most prominent of these is N9b, which is the most frequent mtDNA haplogroup assigned to Jōmon individuals. An estimate of the time to the most recent common ancestor (t MRCA) is ca. 22,000 years ago, which is older than the t MRCA for several other haplogroups found in East Asians (Adachi et al., 2011). The non-recombining region of the Y-chromosome has also been extensively studied in Japan. Y-haplogroup D1b is exceptionally interesting as it is found in high frequency in the Japanese archipelago and the Tibetan Plateau but not elsewhere (Hammer & Horai, 1995; Tajima et al., 2004; Watanabe et al., 2019). The estimated t MRCA for D1b is ca. 19,400 years ago (Hammer et al., 2006), similar to that estimated for N9b mtDNA. These dates, if corresponding to the expansion of Jōmon ancestry on the archipelago, suggest that Jōmon ancestry separated fairly early from mainland East Asian ancestry.
One mtDNA haplogroup found in many Jōmon-associated individuals (e.g. Kanzawa-Kiriyama et al., 2013; Adachi et al., 2011; Kim et al., 2010) as well as Yayoi-associated individuals (Igawa et al., 2009) is D4, which is widespread in northern East Asia both in past and present-day populations (e.g. Tanaka et al., 2004; Zhao et al., 2010; Hong et al., 2014; Jeong et al., 2018; Lan et al., 2019). Its widespread presence, however, suggests that it was found in the common ancestral population of all East Asians, making it difficult to evaluate its role in more recent migrations and interactions.
In genomic studies of ancient Asians, both a 40,000-year-old individual found in Tianyuan Cave, Beijing, China (Fu et al., 2013; Yang et al., 2017) and 8000-year-old individuals from Southeast Asia associated with Hòabìnhian foragers (McColl et al., 2018) show an early split time from other East Asians, including from ancient Jōmon genomes (Kanzawa-Kiriyama et al., 2017, 2019). Thus, we denote them as ‘basal Asians’. In contrast, humans living in Southeast Asia, the eastern steppes of Siberia and the southwest edges of the Tibetan Plateau from the Neolithic to historic periods (e.g. Sikora et al., 2019; Jeong et al., 2016b) share a closer relationship to present-day East Asians than the Jōmon individuals, confirming that Jōmon are distantly related to mainland East Asian ancestry (Yang, MA and Fu, Q unpublished observations).
In fact, the Jōmon sampled thus far probably separated earlier than the split time of East Asians and the early Asian population that contributed to Native Americans (Gakuhari et al., 2019; Kanzawa-Kiriyama et al., 2017, 2019). Estimates of when ancestral Native Americans separated from the East Asian lineage is ca. 36,000–25,000 years ago (Moreno-Mayar et al., 2018), and a direct estimate of the split time between the Jōmon and mainland East Asians ranges from 38,000–18,000 years ago (Kanzawa-Kiriyama et al., 2019). These estimates are broad, indicating there is still high uncertainty as to the exact date. However, studies of both uniparental and nuclear markers all highlight Jōmon ancestry separated from mainland East Asian ancestry in the Palaeolithic, with dates ranging from 38,000–18,000 years ago.
|
|
|
|
Post by Admin on Aug 9, 2021 20:01:42 GMT
Figure 2. F4-tests surveying the relationship of populations with East Asian ancestry (on the x-axis, ‘X’) to the Ikawazu Jōmon, relative to ancient Southeast Asian foragers (Hòabìnhian, HB), an ancient Tibetan (Chokhopani, CH), and an inland ancient northern East Asian (Shamanka_EN, SH) (graphs by Melinda Yang). Numerical values for the data visualized here can be found in Table S3. F4-statistics are tests of symmetry of the form f4(Out, P1; P2, P3), where it is assumed that P2 and P3 form a clade with espect to P1. Our statistics take the form (A) f4(Mbuti, HB; Ikawazu Jōmon, ‘X’ East Asians) or (B, C) f4(Mbuti, Ikawazu Jōmon; CH or SH, ‘X’ East Asians), with the underlying tree structure verified by other genetic analyses. Central African Mbuti were used as the outgroup (Out). 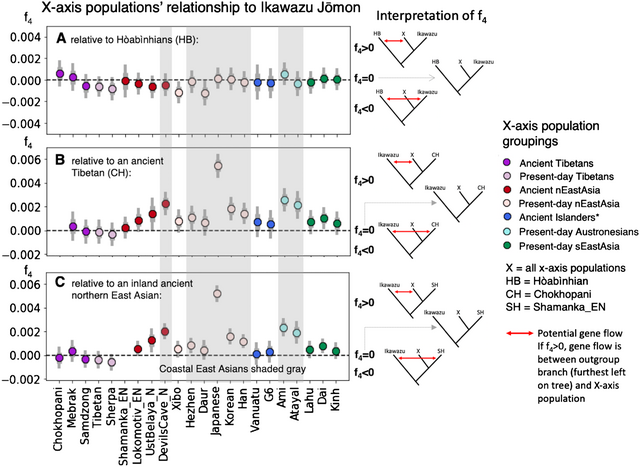 Key: tree models showing relationships of f4 values are placed to the right of the plot: f4 > 0 corresponds to the highest tree; f4 = 0 corresponds to the middle tree (clarified by dashed grey arrow); and f4 < 0 corresponds to the lowest tree for each figure. The presence of gene flow in the model is indicated by the red arrow. A significantly positive value highlights gene flow between the outgroup (P1) and ‘X’ East Asians (P3). A significantly negative value highlights gene flow between the outgroup (P1) and the other population in the tree (P2). Two standard errors are indicated by the thin grey bar, while one standard error is indicated by the thick grey bar. Points are coloured (colour online) by region of origin, with darker colours for ancient populations and lighter colours for present-day populations. East Asians for whom all or part of their population live near coastal East Asia are shaded grey. *Ancient islanders were sampled from Southeast Asian and Southwest Pacific islands, but were previously shown to share a close relationship to present-day Austronesians (McColl et al., 2018; Skoglund et al., 2016). Interpretation: in Figure 2A, 8000-year-old Hòabìnhian (HB) foragers from Laos and Malaysia are an outgroup to the Ikawazu Jōmon individual and present-day and ancient populations of East Asian ancestry. A connection between the Ikawazu Jōmon individual and the Hòabìnhians would result in a significantly negative statistic (f4 < 0), which is not observed. In all cases, the Ikawazu Jōmon shows no connection to Hòabìnhians (f4 ≈ 0). In Figure 2B, we compare against a 3100-year-old Tibetan (Chokhopani, CH), and in Figure 2C, we compare against a 7500-year-old northern East Asian from the eastern steppe region of Siberia (Shamanka_EN, SH). The Ikawazu Jōmon is an outgroup to all East Asians, including these ancient inland East Asians. Thus, a significantly positive statistic (f4 > 0) indicates that the Ikawazu Jōmon shares connections with that East Asian (X) that the ancient inland East Asian Chokhopani or Shamanka_EN do not possess. No connections are observed between the Ikawazu Jōmon and ancient inland East Asians, which would be indicated by a significantly negative statistic (f4 < 0). A symmetric relationship to Ikawazu is observed when comparing with other non-coastal ancient and present-day East Asians (f4 ≈ 0). Coastal East Asians from northern and southern East Asia tend to show significantly positive results (f4 > 0), indicating a connection to the Ikawazu Jōmon that ancient inland East Asians do not share. A key aspect of the observed genome-wide patterns above is that these Jōmon individuals do not share any relationship with sampled ‘basal Asians’. In admixture tests (Patterson et al., 2012), two populations that form a clade (e.g. the Jōmon and mainland present-day East Asians) may sometimes share excess ancestry with an outgroup population that potentially indicates partial shared demographic history, probably via a population closely related to the outgroup. However, Jōmon individuals show no excess connection to ‘basal Asians’, including the Southeast Asian Hòabìnhians relative to present-day East Asians, as indicated by the lack of significantly negative values in the Box (Figure 2A). In particular, comparison of the Southeast Asian Hòabinhians from Laos and Malaysia with mainland East Asians from East Asia and a Jōmon individual (Ikawazu) shows that the East Asians and Jōmon ancestry are equidistant from Hòabìnhian ancestry (Figure 2A), indicating that the Jōmon do not share a special relationship with Hòabìnhians as previously suggested (McColl et al., 2018). Tests of genetic similarity do not show Hòabìnhians or the Jōmon sharing exceptionally high genetic similarity with each other (Figure S1). Shared ancestry between present-day Japanese and recent Southeast Asians are best explained by a common process – gene flow from mainland East Asia, a phenomenon well-characterized by tests of the Dual Structure model in Japan and by observations of gene flow into Southeast Asia (Kanzawa-Kiriyama et al., 2017, 2019; Lipson et al., 2018). In the future, individuals from eastern Eurasia will almost certainly help to clarify how Jōmon ancestry first entered the Japanese archipelago, but the current sampling has not yet shed light on who they might have been. Connections do exist between the Jōmon and East Asians who live along the coast of East Asia. The first Jōmon mtDNA sequenced in 1989, from the Early Jōmon Urawa-I site in the Kantō region (Figure 1), shared a close relationship to present-day Southeast Asians that suggests common ancestry with Southeast Asians (Horai et al., 1989). This haplogroup, called E1a1a, is prevalent in Austronesian-speaking populations of Taiwan and Southeast Asia who share a closer relationship to East Asians than other Southeast Asians. A ca. 8000-year-old individual sampled from Liang Island off the southeast coast of China in Fujian province also shares this haplogroup (Ko et al., 2014). Study of the Liang Island individual's nuclear genome indicates a close genetic relationship to Austronesians (Yang, MA and Fu, Q unpublished observations). Present-day Austronesians, such as the Ami and Atayal from Taiwan (Mallick et al., 2016), share connections with the Ikawazu Jōmon relative to more inland ancient East Asians (Figure 2B and C), potentially indicating admixture between the Jōmon and southern East Asian populations. Comparisons with nuclear genomes from Fujian in southern China show similar connections (Yang, MA and Fu, Q unpublished observations). Thus, rather than Southeast Asian, the primary signal seems to be associated with Austronesians, who derive from an East Asian population associated with southern China. Siberian connections, particularly in Hokkaido, have also been a recurring theme of Jōmon-related genetic studies. Several of the mtDNA haplogroups prevalent in the Japanese and Jōmon but rare in mainland East Asians, especially G1b, can be found in southeastern Siberians, highlighting potential contacts along the coast in northern East Asia as well (Sato et al., 2009; Adachi et al., 2011). Pre-modern Ainu show similar mtDNA haplogroups to populations in the Lower Amur region of Siberia and populations belonging to the early historic Okhotsk culture of Hokkaido (600–1200 AD) (Adachi, Kakuda, Takahashi, Kanzawa-Kiriyama, & Shinoda, 2018). Studies of nuclear DNA from Early Neolithic populations in the Primorye region of Russia highlight that the Jōmon are an outgroup to all mainland East Asians, including these more northern populations in Siberia (Sikora et al., 2019; de Barros Damgaard et al., 2018). In the same comparisons set up for coastal southern East Asians above, it can be shown that ancient and present-day coastal northern mainland East Asians dating up to 7700 years ago share connections with the Ikawazu Jōmon relative to more inland East Asians (Figure 2B, C). Ancient northern Siberian ancestry prevalent during the Palaeolithic notable for both its closer relationship with European-related rather than Asian-related ancestry and its impact on Native American ancestry (Raghavan et al., 2014; Sikora et al., 2019) is not found in mainland East Asians or the Jōmon, which emphasizes that the connections are specific to coastal mainland East Asians and the Jōmon. |
|





