|
|
Post by Admin on Sept 18, 2021 22:58:47 GMT
 Ancient DNA extracted from human bones has rewritten early Japanese history by underlining that modern day populations in Japan have a tripartite genetic origin—a finding that refines previously accepted views of a dual genomic ancestry. Twelve newly sequenced ancient Japanese genomes show that modern day populations do indeed show the genetic signatures of early indigenous Jomon hunter-gatherer-fishers and immigrant Yayoi farmers—but also add a third genetic component that is linked to the Kofun peoples, whose culture spread in Japan between the 3rd and 7th centuries. Rapid cultural transformations The Japanese archipelago has been occupied by humans for at least 38,000 years but Japan underwent rapid transformations only in the last 3,000 years, first from foraging to wet-rice farming, and then to a technologically advanced imperial state. The previous, long-standing hypothesis suggested that mainland Japanese populations derive dual-ancestry from the indigenous Jomon hunter-gatherer-fishers, who inhabited the Japanese archipelago from around 16,000 to 3,000 years ago, and later Yayoi farmers, who migrated from the Asian continent and lived in Japan from around 900 BC to 300 AD. But the 12 newly sequenced ancient Japanese genomes—which came from the bones of people living in pre- and post-farming periods—also identify a later influx of East Asian ancestry during the imperial Kofun period, which lasted from around 300 to 700 AD and which saw the emergence of political centralisation in Japan.  Shigeki Nakagome, Assistant Professor in Psychiatry in Trinity College Dublin's School of Medicine, led the research, which brought together an interdisciplinary team of researchers from Japan and Ireland. Professor Nakagome said: "Researchers have been learning more and more about the cultures of the Jomon, Yayoi, and Kofun periods as more and more ancient artefacts show up, but before our research we knew relatively little about the genetic origins and impact of the agricultural transition and later state-formation phase." "We now know that the ancestors derived from each of the foraging, agrarian, and state-formation phases made a significant contribution to the formation of Japanese populations today. In short, we have an entirely new tripartite model of Japanese genomic origins—instead of the dual-ancestry model that has been held for a significant time." Genomic insights into key Japanese transformations In addition to the overarching discovery, the analyses also found that the Jomon maintained a small effective population size of around 1,000 over several millennia, with a deep divergence from continental populations dated to 20,000-15,000 years ago—a period which saw Japan become more geographically insular through rising sea-levels. The Japanese archipelago had become accessible through the Korean Peninsula at the beginning of the Last Glacial Maximum, some 28,000 years ago, enabling movement between. And the widening of the Korea Strait 16,000 to 17,000 years ago due to rising sea-levels may have led to the subsequent isolation of the Jomon lineage from the rest of the continent. These time frames also coincide with the oldest evidence of Jomon pottery production.  "The indigenous Jomon people had their own unique lifestyle and culture within Japan for thousands of years prior to the adoption of rice farming during the subsequent Yayoi period. Our analysis clearly finds them to be a genetically distinct population with an unusually high affinity between all sampled individuals—even those differing by thousands of years in age and excavated from sites on different islands," explained Niall Cooke, Ph.D. Researcher at Trinity. "These results strongly suggest a prolonged period of isolation from the rest of the continent." The spread of agriculture is often marked by population replacement, as documented in the Neolithic transition throughout most of Europe, with only minimal contributions from hunter-gatherer populations observed in many regions. However, the researchers found genetic evidence that the agricultural transition in prehistoric Japan involved the process of assimilation, rather than replacement, with almost equal genetic contributions from the indigenous Jomon and new immigrants associated with wet-rice farming. Several lines of archaeological evidence support the introduction of new large settlements to Japan, most likely from the southern Korean peninsula, during the Yayoi-Kofun transition. And the analyses provide strong support for the genetic exchange involved in the appearance of new social, cultural, and political traits in this state-formation phase. "The Japanese archipelago is an especially interesting part of the world to investigate using a time series of ancient samples given its exceptional prehistory of long-standing continuity followed by rapid cultural transformations. Our insights into the complex origins of modern-day Japanese once again shows the power of ancient genomics to uncover new information about human prehistory that could not be seen otherwise," added Dan Bradley, Professor of Population Genetics in Trinity's School of Genetics and Microbiology, who co-led the project. The eye-opening research has just been published in Science Advances. Explore further New genetic evidence resolves origins of modern Japanese More information: "Ancient genomics reveals tripartite origins of Japanese populations," Science Advances (2021). www.science.org/doi/10.1126/sciadv.abh2419Journal information: Science Advances |
|
|
|
Post by Admin on Sept 19, 2021 3:58:34 GMT
Ancient genomics reveals tripartite origins of Japanese populations
Abstract
Prehistoric Japan underwent rapid transformations in the past 3000 years, first from foraging to wet rice farming and then to state formation. A long-standing hypothesis posits that mainland Japanese populations derive dual ancestry from indigenous Jomon hunter-gatherer-fishers and succeeding Yayoi farmers. However, the genomic impact of agricultural migration and subsequent sociocultural changes remains unclear. We report 12 ancient Japanese genomes from pre- and postfarming periods. Our analysis finds that the Jomon maintained a small effective population size of ~1000 over several millennia, with a deep divergence from continental populations dated to 20,000 to 15,000 years ago, a period that saw the insularization of Japan through rising sea levels. Rice cultivation was introduced by people with Northeast Asian ancestry. Unexpectedly, we identify a later influx of East Asian ancestry during the imperial Kofun period. These three ancestral components continue to characterize present-day populations, supporting a tripartite model of Japanese genomic origins.
INTRODUCTION
The Japanese archipelago has been occupied by humans for at least 38,000 years. However, its most radical cultural transformations have only occurred within the past 3000 years, during which time its inhabitants quickly transitioned from foraging to widespread rice farming to a technologically advanced imperial state (1, 2). These rapid changes, coupled with geographical isolation from continental Eurasia, make Japan a unique microcosm in which to study the migratory patterns that accompanied agricultural spread and economic intensification in Asia. Before the arrival of farming cultures, the archipelago was occupied by diverse hunter-gatherer-fisher groups belonging to the Jomon culture, characterized by their use of pottery. The Jomon period began during the Oldest Dryas that followed the Last Glacial Maximum (LGM) (3), with the earliest pottery shards dating to ~16,500 years ago (ka ago), making these populations some of the oldest users of ceramics in the world (2). Jomon subsistence strategies varied and population densities fluctuated through space and time (4), with trends toward sedentism. This culture continued until the beginning of the Yayoi period (~3 ka ago), when the arrival of paddy field rice cultivation led to an agricultural revolution in the archipelago. This was followed by the Kofun period, starting ~1.7 ka ago, which saw the emergence of political centralization and the imperial reign that came to define the region (1).
An enduring hypothesis on the origin of modern Japanese populations proposes a dual-structure model (5), in which Japanese populations are the admixed descendants of the indigenous Jomon and later arrivals from the East Eurasian continent during the Yayoi period. This hypothesis was originally proposed on the basis of morphological data but has been widely tested and evaluated across disciplines [see a recent review in (6)]. Genetic studies have identified population stratifications within present-day Japanese populations, supporting at least two waves of migrations to the Japanese archipelago (7–10). Previous ancient DNA studies have also illustrated the genetic affinity of Jomon and Yayoi individuals to Japanese populations today (11–15). Still, the demographic origins and impact of the agricultural transition and later state formation phase are largely unknown. From a historical linguistic standpoint, the arrival of proto-Japonic language is theorized to map to the development of Yayoi culture and the spread of wet rice cultivation (6). However, archaeological contexts and their continental affiliations are distinct between the Yayoi and Kofun periods (1); whether the spread of knowledge and technology was accompanied by major genetic exchange remains elusive.
Here, we report 12 newly sequenced ancient Japanese genomes spanning 8000 years of the archipelago’s pre- and protohistory (Fig. 1 and Table 1). To our knowledge, this is the largest set of time-stamped genomes from the archipelago, including the oldest Jomon individual and the first genomic data from the imperial Kofun period. We also include five published prehistoric Japanese genomes in our analysis: three Jomon individuals (F5 and F23 from the Late Jomon period and IK002 from the Final Jomon period) (12–14), as well as two 2000-year-old individuals associated with the Yayoi culture from the northwestern part of Kyushu Island, where skeletal remains exhibit Jomon-like characters rather than immigrant types but other archaeological materials clearly support their association with the Yayoi culture (15, 16). Despite this morphological assessment (16), these two Yayoi individuals show an increased genetic affinity to present-day Japanese populations compared with the Jomon, implying that admixture with continental groups was already advanced by the Late Yayoi period (15). Integrating these Japanese genomes with a larger ancient genomic dataset spanning the Central and Eastern Steppe (17, 18), Siberia (19), Southeast Asia (12), and East Asia (15, 20, 21), our study aims to better characterize the preagricultural populations of the Jomon period, as well as the subsequent migrations and admixtures that have shaped the genetic profile of the archipelago today.
|
|
|
|
Post by Admin on Sept 19, 2021 21:30:19 GMT
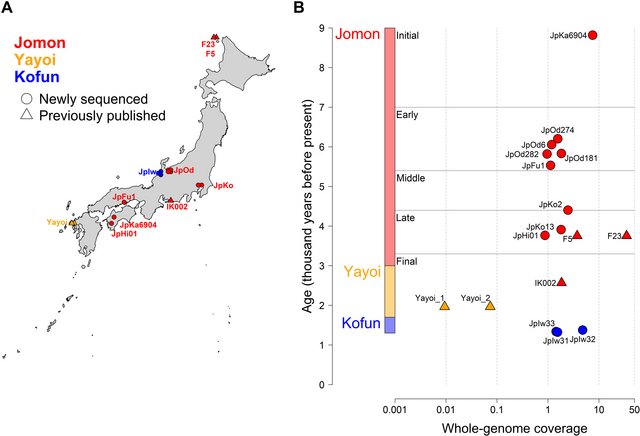 Fig. 1. Sampling locations, dates, and genome coverage of ancient Japanese individuals. (A) Archaeological sites are marked with circles for individual genomes newly sequenced in this study and triangles if previously reported (see Table 1 and table S1). The colors represent three different periods of Japanese pre- and protohistory: Jomon, Yayoi, and Kofun. (B) Each individual is plotted with whole-genome coverage on the x axis and median age (years before present) on the y axis. The nine Jomon individuals are split into five different subperiods on the basis of their ages (see note S1): Initial (JpKa6904), Early (JpOd274, JpOd6, JpOd282, JpOd181, and JpFu1), Middle (JpKo2), Late (JpKo13, JpHi01, F23, and F5), and Final (IK002).
ASSOCIATED CULTURE SAMPLE ID DATE RANGE AND MEDIAN (CAL B.P.) COVERAGE MTDNA CONTAMINATION RATE(%) MOLECULAR SEX MTDNA YDNA HAPLOGROUP REF.
NEWLY SEQUENCED IN THIS STUDY
JOMON JpKa6904 8646–8991; 8819 7.51 1.46 XX N9b3 – –
JPOD274 6119–6289; 6204 1.56 1.13 XY M7a D1b1d1 –
JPOD6 5934–6179; 6057 1.18 1.55 XX N9b3 – –
JPOD181 5751–5917; 5834 1.83 0.91 XY N9b1 D1b1d1 –
JPOD282 5737–5902; 5820 0.96 1.38 XY M7a1 D1b1d1 –
JPFU1 5478–5590; 5534 1.13 2.15 XX M7a1 – –
JPKO2 4294–4514; 4404 2.47 1.44 XX N9b – –
JPKO13 3847–3978; 3913 1.81 1.50 XX N9b1 – –
JPHI01 3685–3850; 3768 0.88 1.45 XX M7a1a – –
KOFUN JpIw32 1347–1409; 1378 4.80 0.41 XY B5a2a1b O3a2c –
JPIW31 1303–1377; 1340 1.44 0.63 XX D5c1a – –
JPIW33 1295–1355; 1325 1.54 0.75 XX M7b1a1a1 – –
PREVIOUSLY PUBLISHED
JOMON F23 3550–3960; 3755 34.82 1.20 XX N9b1 - (14)
F5 – 3.74 2.45 XY N9b1 D1b2b (14)
IK002 2418–2720; 2569 1.85 0.50 XX N9b1 – (12)
YAYOI Yayoi_1 – 0.01 2.92 XX M7a1a4 – (15)
YAYOI_2 1931–2001; 1966 0.07 2.33 XY D4a1 O (15)
Table 1. Summary of ancient Japanese data.
|
|
|
|
Post by Admin on Sept 20, 2021 1:57:31 GMT
RESULTS
A time series of ancient genomes from pre- and protohistoric Japan
Our initial screening focused on 14 ancient skeletal remains excavated from six archaeological sites across the archipelago (see note S1). High levels of endogenous human DNA were preserved in 12 of these samples (Table 1), which were then further shotgun-sequenced to higher coverage, ranging from 0.88× to 7.51× (Fig. 1 and table S1). Nine of the 12 samples are associated with the Jomon culture, representing the west and central parts of the archipelago and four different stages of the Jomon period (Initial, Early, Middle, and Late Jomon) (Fig. 1). The remaining three samples date to ~1.3 ka ago, falling within the Kofun period. We confirm that all newly sequenced genomes show postmortem damage patterns (fig. S1) and a low level of modern human contamination (<2.15%) (Table 1 and table S2). Our kinship analysis confirms that all pairs of individuals are unrelated (fig. S2). Mitochondrial haplogroups for all Jomon individuals belong to the N9b or M7a clades, which are strongly associated with this population (11–14, 22) and rare outside of Japan today (23). The three Jomon males (table S3) belong to the Y chromosome haplogroup D1b1, which is present in modern Japanese populations but almost absent in other East Asians (24). In contrast, the Kofun individuals all belong to mitochondrial haplogroups that are common in present-day East Asians (25), while the single Kofun male has the O3a2c Y chromosome haplogroup, which is also found throughout East Asia, particularly in mainland China (26). To place our data within the wider context of East Eurasian demography, we combined the ancient Japanese genomes with genomic data from previously published ancient (table S4 and fig. S3) and present-day individuals. Throughout this study, the modern Japanese population is represented by either data from the Simons Genome Diversity Project (SGDP) (27) or JPT (i.e., Japanese in Tokyo) in the 1000 Genome Project phase 3 (28). However, we note that ancestral heterogeneity exists across the archipelago today, which is not fully captured by this standard reference set. Other ancient and present-day populations analyzed in this study are primarily labeled by either geographic or cultural contexts.
Genetic distinction between different cultural periods
We explored the genetic diversity within our time series data by looking at the shared genetic drift between all pairwise comparisons of individuals from both the ancient and modern (SGDP) Japanese populations using the statistic f3(Individual_1, Individual_2; Mbuti) (Fig. 2A). Our results very clearly define three distinct clusters of Jomon, Yayoi, and Kofun individuals, the last of whom group with the modern Japanese individuals, suggesting that cultural shifts were accompanied by genomic changes. Despite the large spatial and temporal variation in the Jomon dataset, extremely high levels of shared drift are observed between all 12 individuals. The Yayoi individuals are most closely related to each other and also have a higher affinity to the Jomon than to the Kofun individuals. The Kofun and modern Japanese individuals are almost indistinguishable from one another by this metric, implying some level of genetic continuity over the past 1400 years.
|
|
|
|
Post by Admin on Sept 20, 2021 4:43:29 GMT
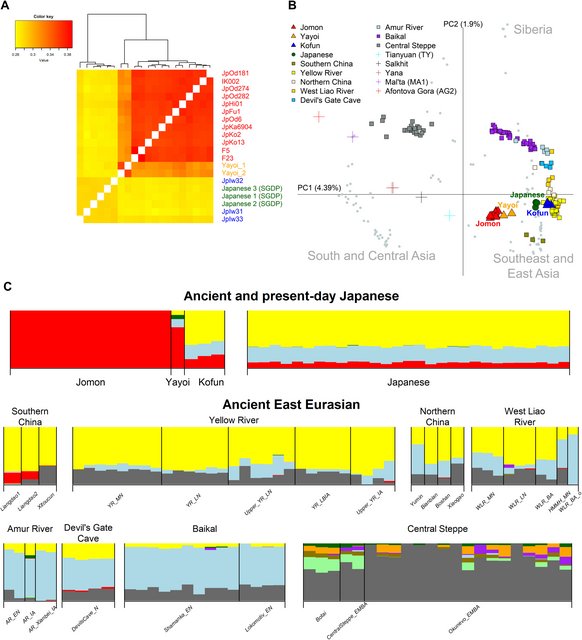 Fig. 2. Genetic diversity through time in Japan. (A) A heatmap of pairwise outgroup f3 statistic comparisons between all ancient and modern Japanese individuals. (B) Principal components analysis (PCA) visualizing ancient Japanese individuals (i.e., Jomon, Yayoi, and Kofun) and continental ancient individuals (presented as colored symbols) projected onto 112 present-day East Eurasians (gray circles with Japanese highlighted in dark green). (C) Selected individuals from an ADMIXTURE analysis (K = 11; complete pictures from K = 2 to K = 12 are presented in figs. S5 and S6), showing a distinct Jomon ancestral component (represented by red), a component common in ancient samples from the Baikal region and the Amur River basin (represented by light blue) and a broad East Asian component (represented by yellow). A gray component that is most dominant in the Central Steppe is absent in the ancient and modern Japanese samples. The middle and bottom rows show selected Ancient East Eurasian populations from each geographical regions: Southern China (from left to right: Liangdao1, Liangdao2, and Xitoucun), Yellow River (YR) (Middle Neolithic, YR_MN; Late Neolithic, YR_LN; Late Neolithic, Upper_YR_LN; Late Bronze Age/Iron Age, YR_LBIA; and Iron Age, Upper_YR_IA), Northern China (Yumin, Bianbian, Boshan, and Xiaogao), West Liao River (WLR) (Middle Neolithic, WLR_MN; Late Neolithic, WLR_LN; Bronze Age, WLR_BA; Middle Neolithic individual from Haminmangha, HMMH_MN; and Bronze Age individual with a different genetic background from WLR_BA and WLR_BA_o), Amur River (AR) (Early Neolithic, AR_EN; and Iron Age, AR_IA and AR_Xianbei_IA), Baikal (Early Neolithic, Shamanka_EN and Lokomotiv_EN), and Central Steppe (Botai, CentralSteppe_EMBA, and Okunevo_EMBA). We further investigated the genome-wide autosomal affinities of our ancient Japanese individuals to continental populations using principal components analysis (PCA). We projected ancient individuals onto the genetic variation of present-day populations in the SGDP dataset from South and Central Asia, Southeast and East Asia, and Siberia (Fig. 2B and fig. S4). We observe that the ancient Japanese individuals separate into their respective cultural designations along PC1. All Jomon individuals form a tight cluster, falling apart from other ancient populations, as well as present-day Southeast and East Asians, suggesting a sustained geographic isolation. The two Yayoi individuals appear near to this Jomon cluster, supporting genetic and morphological similarity to Jomon as reported in (15, 16). However, a shift toward East Asian populations implies the presence of additional continental ancestry in Yayoi. Ancient individuals from East Eurasia show a geographic cline from the south to the north on PC2: Southern China, Yellow River, Northern China, West Liao River, Devil’s Gate Cave, Amur River, and Baikal. The three individuals from the Kofun period fall within the diversity of the Yellow River cluster. ADMIXTURE analysis with the Human Origins Array dataset also supports the increase in continental gene flow into the archipelago after the end of the Jomon period (Fig. 2C and figs. S5 and S6). The Jomon have a distinct ancestral component (represented in red in Fig. 2C), which is also visible at high levels in the Yayoi and remains at reduced levels in the Kofun and Japanese. New ancestral components appear in the Yayoi, at proportions similar to the profile seen in the Amur River basin and surrounding regions. These include a larger component that is dominant in Northeast Asians (represented by light blue) and another smaller component that represents much broader East Asian ancestry (represented by yellow). This East Asian component becomes dominant in the Kofun period and the modern Japanese population. Deep divergence of the Jomon lineage by geographic isolation The separation of the Jomon from other populations (Fig. 2) supports the idea that they form a distinct lineage among East Eurasians, as proposed in previous studies (13, 14). To explore the depth of this divergence, we reconstructed the phylogenetic relationship of Jomon with 17 other ancient and present-day populations using TreeMix with differing numbers of admixture events (Fig. 3A and fig. S7) (29). Our results infer that Jomon emerged after the early divergences of Upper Paleolithic East Eurasians (Tianyuan and Salkhit) and ancient Southeast Asian hunter-gatherers (Hoabinhian), but before the splitting off of other samples including present-day East Asians, an ancient Nepali (Chokhopani), hunter-gatherers from Baikal (Shamanka_EN and Lokomotiv_EN) and Chertovy Vorota Cave (Devil’s Gate Cave) in the Primorye Region, and a Pleistocene Alaskan (USR1). We further confirm the position of Jomon between two other deeply diverged hunter-gatherer lineages in this tree through formal tests of symmetric models using f4(Mbuti, X; Hoabinhian/DevilsCave_N, Jomon) (fig. S8, A and B). These show that all East Asian individuals in our dataset since the beginning of the Jomon period have a higher affinity to Jomon than the earlier diverged Hoabinhian but have a lower affinity in comparison to DevilsCave_N; this supports the inferred phylogeny of three distinct hunter-gatherer lineages in East Eurasia, rather than a previously proposed model in which Jomon is a mixture of Hoabinhian and East Asian-related lineages [f3(Jomon; Hoabinhian, DevilsCave_N) = 0.193, Z = 61.355; table S5] (12, 30). We also consistently infer gene flow from Jomon to the modern Japanese across all migration models tested, with genetic contributions ranging from 8.9 to 11.5% (fig. S7). This is consistent with the mean Jomon component of 9.31% in the present-day Japanese individuals estimated from our ADMIXTURE analysis (Fig. 2C). These results suggest a deep divergence of the Jomon and an ancestral link to present-day Japanese populations. |
|





