|
|
Post by Admin on Sept 20, 2021 19:53:09 GMT
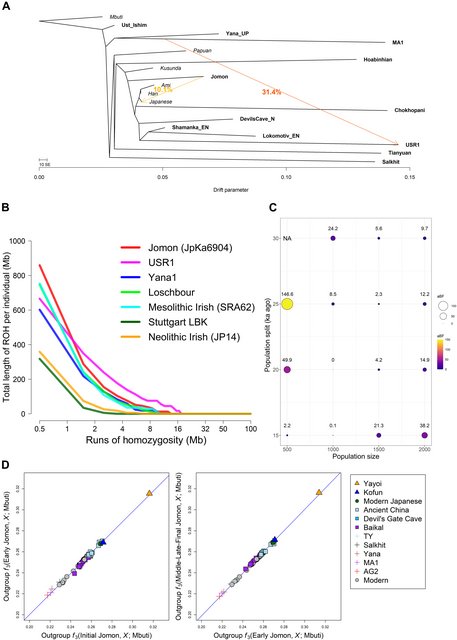 Fig. 3. Demographic history of the Jomon lineage. (A) Maximum likelihood phylogenetic tree reconstructed by TreeMix under a model of two migrations. The tree shows a phylogenetic relationship among ancient (bold) and present-day (italic) populations. Colored arrows represent the migration pathways. The migration weight represents the fraction of ancestry derived from the migration edge. All other migration models from m = 0 to m = 5 are presented in fig. S7. (B) ROH spectra for Mesolithic and Neolithic hunter-gatherers including the 8.8-ka-old JpKa6904. Total length of ROH is plotted against different sizes of homozygous fragments ranging from 0.5 to 100 Mb. (C) Fitting of the models under different combinations of N (x axis) and T (y axis) for the 8.8-ka-old Jomon individual. Each point in the balloon plot represents a log10-scaled approximate Bayes factor (aBF) that compares likelihoods between a model with the highest likelihood and each of the other given models; the point with aBF = 0 is the model with the highest likelihood (N = 1000 and T = 20 ka ago; see fig. S10). NA means that aBF is not measurable for the model due to its likelihood equal to zero. (D) A comparison of outgroup f3 statistic results for the Jomon dataset divided into three subperiods measured using f3(Jomon_Sub-Period, X; Mbuti) (see fig. S12 for an expanded analysis). The three subperiods are as follows: Initial Jomon (JpKa6906); Early Jomon (JpFu1, JpOd6, JpOd181, JpOd274, and JpOd282); and a merged group for all Middle, Late, and Final Jomon (F5, F23, IK002, JpHi01, JpKo2, and JpKo13). We apply population genetic modeling to estimate the timing of the appearance of the Jomon lineage. Our approach makes use of genome-wide patterns of runs of homozygosity (ROHs) to identify the demographic scenario that best fits the ROH spectrum observed in our oldest and highest coverage sample, JpKa6904 (see note S2). The distribution of ROH tracts reflects the effective population size and the time to the most recent common ancestor between two copies of haplotypes within an individual (31, 32). The 8.8-ka-old Jomon carries high levels of ROH, in particular with the highest frequency of short ROH (due to population effects rather than recent inbreeding) yet reported (Fig. 3B) (33). This pattern, coupled with strong shared genetic drift among the Jomon individuals (fig. S9), implies that the Jomon population underwent a severe population bottleneck. With a search of parameter space of population size and split time, our estimates place the appearance of the Jomon lineage between 15 and 20 ka ago, followed by the maintenance of a very small population size of ~1000 until at least the Initial Jomon period (Fig. 3C and figs. S10 and S11). This coincides with rising sea levels and the severing of the land bridge to the mainland at the end of the LGM (34, 35) and shortly precedes the first appearance of Jomon pottery in the archipelago (2). We then asked whether the Jomon had any contact with continental Upper Paleolithic people after the divergence of their lineage, but before their isolation in the archipelago, using the statistic f4(Mbuti, X; Jomon, Han/Dai/Japanese) (fig. S8, C to E). Among the Upper Paleolithic individuals tested, only Yana_UP is significantly closer to Jomon than Han, Dai, or Japanese, respectively (Z > 3.366). This affinity is still detectable even if we replace these reference populations with the other Southeast and East Asians (table S6), supporting gene flow between the ancestors of Jomon and Ancient North Siberians, a population widespread in North Eurasia before the LGM (19). We lastly investigated potential temporal and spatial variation within the Jomon population. Three temporal groups, defined by the Initial, Early, and Middle-Late-Final stages of the Jomon period, show similar levels of shared genetic drift with ancient and present-day continental populations, implying little or no genetic impact from outside of the archipelago over these subperiods (Fig. 3D and fig. S12). This pattern is further backed up by the absence of any significant gene flow, observed with the statistic f4(Mbuti, X; sub_Jomoni, sub_Jomonj), where i and j are any pairs of the three Jomon groups (fig. S13). These Jomon individuals similarly do not display any diversity in genetic affinity to continental populations when grouped by geography (i.e., different islands on which the samples are located: Honshu, Shikoku, and Rebun Island) (fig. S14). The only observable difference within the Jomon individuals is a slightly higher affinity between sites located on Honshu, implying an insular effect with restricted gene flow between Honshu and other islands (fig. S15). Overall, these results show limited spatial and temporal genetic variation within the Jomon population, supporting the idea of near-complete isolation from the rest of Asia for thousands of years. |
|
|
|
Post by Admin on Sept 20, 2021 22:19:27 GMT
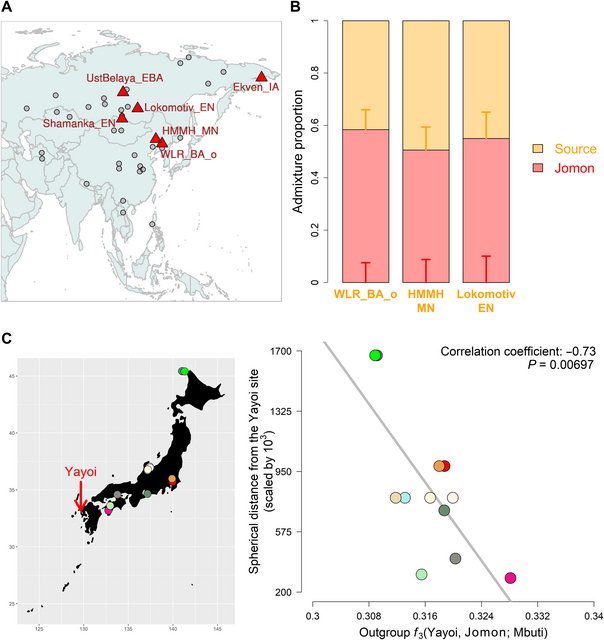 Fig. 4. Genetic changes in the Yayoi period. (A) Geographical map highlighting results from f4(Mbuti, X; Jomon, Yayoi); continental ancient populations who are significantly closer to the Yayoi than the Jomon (with Z > 3.0) are represented by red triangles, while those who are symmetrically related to the both populations are represented by gray circles. (B) Genetic ancestry of the Yayoi modeled with a two-way admixture of the Jomon and the other source represented by: Bronze Age or Middle Neolithic individuals from West Liao River (WLR_BA_o or HMMH_MN) or Baikal hunter-gatherer (Lokomotiv_EN). Vertical bars represent ±1 SE estimated by qpAdm. The values of admixture proportions are shown in table S7. (C) Correlation of shared genetic drift between the Yayoi and a Jomon individual with the geographic locations of Jomon sites. The spherical distance from the Yayoi site is measured with the Haversine formula (93). The map marks the archaeological site of Yayoi with a red arrow and the sites of Jomon by circles with different colors. To further distinguish these six potential sources of continental ancestry, we modeled the Yayoi as a two-way admixture of the Jomon and each in turn using qpWave (table S7). The admixture model was confidently supported (P > 0.05) for three of these: Baikal hunter-gatherers and the West Liao Middle Neolithic or Bronze Age individuals with a high level of Amur River ancestry (20). These groups all share a dominant Northeast Asian ancestral component (Fig. 2C and fig. S17) (20). Admixture fractions of 55.0 ± 10.1%, 50.6 ± 8.8%, or 58.4 ± 7.6% for the Jomon input were estimated by qpAdm when each of these three were, respectively, used as the second source (Fig. 4B and table S7), with a fraction of 61.3 ± 7.4% returned when the West Liao Middle Neolithic and Bronze Age individuals were merged into a single source population (table S7). We further confirm by f4(Mbuti, Jomon; Yayoi_1, Yayoi_2) that the level of Jomon ancestry is comparable between these two Yayoi individuals (Z = 1.309). These results imply an approximately equal ratio of indigenous hunter-gatherer and migrant contribution to the Yayoi communities associated with this northwestern Kyushu site. This parity is particularly notable when compared to agricultural migrations in western Eurasia, where minimal hunter-gatherer contribution is observed in many regions, including the archipelago of Britain and Ireland (37–40), which mirrors Japan as an insular geographic extreme of Eurasia. While the West Liao populations used in our admixture models did not themselves practice rice farming, they are situated just north of a hypothesized route of agricultural spread into Japan, to which our results lend weight. This follows the Shandong Peninsula (northeastern China) into the Liaodong Peninsula (northwestern part of the Korean Peninsula) and then reaches the archipelago via the Korean Peninsula (41). We further investigated how the Yayoi culture spread into the archipelago using outgroup f3 statistics that measured genetic affinity between the Yayoi and each of the Jomon individuals. We find that the strength of shared genetic drift has a significant correlation with the distance from the location of the Yayoi individuals (P = 0.00697; Fig. 4C); the closer the Jomon archaeological sites are to the Yayoi site, the more the Jomon individuals share genetic drift with the Yayoi. This result supports the introduction of rice farming via the Korean Peninsula (41, 42), followed by admixture with local Jomon populations in the south of the archipelago. |
|
|
|
Post by Admin on Sept 21, 2021 0:59:09 GMT
Genetic ancestry of migrants during the Kofun period Historical records provide strong support for continued population movement from the continent to the archipelago during the Kofun period (1). However, our qpWave modeling of the three Kofun individuals rejected the two-way admixture of Jomon and Northeast Asian ancestry that fitted the Yayoi individuals (P < 0.05; table S8). Thus, the Kofun are genetically distinct from the Yayoi in terms of their ancestral components, as supported from our outgroup f3, PCA, and ADMIXTURE clusters (Fig. 2), as well as from previous morphological studies (43, 44). To identify additional ancestral groups that contributed to the genetic makeup of the Kofun individuals, we tested genetic affinity between the Kofun and each of the continental populations using f4(Mbuti, X; Yayoi, Kofun) (Fig. 5A and fig. S18). We find that most of the ancient or modern populations in our dataset are significantly closer to the Kofun than they are to the Yayoi. This finding implies additional migration to the archipelago during the six centuries that separates the genomes from these two periods. 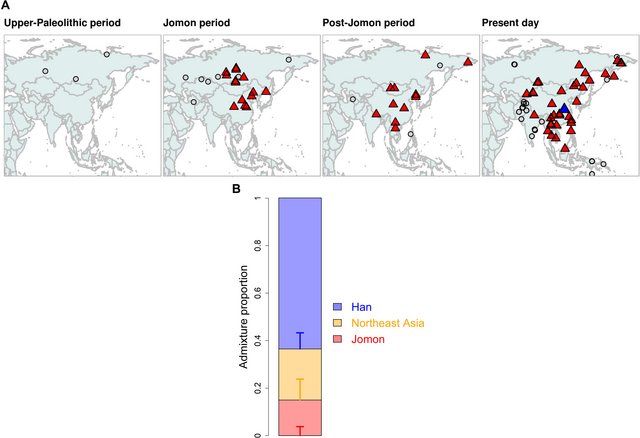 Fig. 5. Genetic changes in the Kofun period. (A) Geographical maps highlighting results from f4(Mbuti, X; Yayoi, Kofun); continental ancient and present-day populations who are significantly closer to the Kofun than the Yayoi with a Z score of >3.0 are represented by red triangles, while those that are symmetrically related to both populations are represented by gray circles. The populations tested are split into four different periods, depending on their ages: Upper-Paleolithic (>16,000 years B.P.), Jomon (from 16,000 to 3000 years B.P.), Post-Jomon (from 3000 years B.P. to the present), and present day. Han is highlighted by a blue triangle in the present-day panel. (B) Genetic ancestry of the Kofun individuals modeled with a three-way admixture of the Jomon, Northeast Asia (WLR_BA_o and HMMH_MN), and Han. Vertical bars represent ±1 SE estimated by qpAdm. The values of admixture proportions are shown in table S10. We attempted to narrow the source of this migration by testing the fit of two-way admixtures of the Yayoi and populations identified from our f4 statistics as being significantly closer to the Kofun (table S9). This admixture model was confidently supported with P > 0.05 for only 5 of 59 populations tested. We then applied qpAdm to quantify the genetic contribution from Yayoi and each of these sources in turn (table S9). The two-way admixture model was subsequently rejected in an additional two populations due to a lack of support from different reference sets. The remaining three populations (Han, Korean, and YR_LBIA) show 20 to 30% contributions to the Kofun (fig. S19). These three all share strong genetic drift (highlighted by the green square in fig. S17), characterized with a major component of broadly East Asian ancestry in their ADMIXTURE profiles (fig. S6). To further screen the source of additional ancestry in the Kofun individuals, we tested a three-way admixture by replacing the Yayoi ancestry with Jomon and Northeast Asian ancestry (table S10). Only Han were successfully modeled as a source of ancestry in the model (Fig. 5B), with a significantly better fitting of the three-way admixture than any possible two-way admixture models (table S11). Given that Jomon ancestry is diluted by approximately a factor of 4 between the Yayoi and Kofun populations sampled, these results suggest that the state formation phase saw a large influx of migrants who had East Asian ancestry. We then explore the possibility that the continental ancestry observed in both the Yayoi and Kofun periods derives from the same source, with intermediate levels of Northeast and East Asian ancestry (table S12). Only one candidate was found to better fit a two-way mixture for Kofun, a population of the Late Bronze Age and Iron Age individuals from the Yellow River basin (YR_LBIA) (20), although this was not consistent across the reference sets (nested, P = 0.100; table S13). Despite not showing statistically significant gene flow with Yayoi to the exclusion of Jomon (Z = 2.487) (Fig. 4A and fig. S16), we find that a two-way model between YR_LBIA and Jomon also fits the Yayoi (table S14). This Yellow River population has an intermediate genetic profile with approximately 40% Northeast Asian and 60% East Asian (i.e., Han) ancestry, as estimated by qpAdm. Thus, this is an intermediate genetic profile that fits both Yayoi and Kofun in certain models, with an input of 37.4 ± 1.9% and 87.5 ± 0.8% in these populations (table S14). These results imply that continuous gene flow from a single source may be sufficient to explain the genetic changes between the Yayoi and Kofun. However, our broader analyses strongly suggest that a single source of gene flow is less likely than two distinct waves of migrations. First, the ratios of Northeast Asian to East Asian ancestry identified in ADMIXTURE were starkly different between the Yayoi (1.9:1) and the Kofun (1:2.5) (Fig. 2C). Second, this contrast in continental affinity is also observable in the different forms of f statistics, in which there is a repeated pattern of Yayoi having significant affinity to those with Northeast Asian ancestry (Fig. 4A and fig. S16), while the Kofun individuals form a tight cluster with other East Asians including Han and ancient Yellow River populations (Fig. 5A and figs. S17 and S18). Last, we find support for a two-pulse model from our dating of the admixture in the Kofun individuals by DATES (fig. S20) (45). A single admixture event with the intermediate population (i.e., YR_LBIA) is estimated to have occurred 1840 ± 213 years before the present (B.P.), which is much later than the onset of the Yayoi period (~3 ka ago). In contrast, if two separate admixture events with two distinct sources are assumed, the resulting estimates reasonably fit the timings consistent with the beginning of the Yayoi and Kofun periods (3448 ± 825 years B.P. for the admixture between Jomon and Northeast Asian ancestry and 1748 ± 175 years B.P. for Jomon and East Asian ancestry; fig. S20). These genetic findings are further supported by both the archaeological evidence and the historical records, which document the arrival of new people from the continent during the period (1). Genetic heritage of Kofun in present-day Japanese The three Kofun individuals are genetically similar to the present-day Japanese, as shown in Fig. 2. This implies no substantial change in the genetic makeup of Japanese populations since the Kofun period. To look for signals of additional genetic ancestry in the present-day Japanese samples, we tested whether the continental populations have preferential affinity to modern genomes relative to Kofun using f4(Mbuti, X; Kofun, Japanese) (fig. S21). Although some of the ancient populations exhibit higher affinity to Kofun than Japanese, none of them is supported by qpAdm as an additional source of ancestry present in the Kofun (see note S3 and table S15). Unexpectedly, no ancient or modern population shows additional gene flow with the present-day Japanese to the exclusion of the Kofun. Our admixture modeling further confirms that the present-day Japanese population is sufficiently explained by Kofun ancestry without increasing Jomon or Yayoi ancestry or without introducing an additional ancestor represented by present-day Southeast or East Asians or Siberians (table S16). We also find that the modern Japanese population has the same set of ancestral components as the three-way admixture in the Kofun individuals (table S17), with a slightly increased level of East Asian ancestry in the modern Japanese compared with the Kofun individuals sampled (fig. S22). This suggests a certain level of genetic continuity but not absolute. A strict model of continuity between the Kofun and the modern Japanese population (i.e., with no genetic drift unique to the Kofun lineage) is rejected (table S18) (46). However, we note no dilution of Jomon ancestry in the Japanese population (15.0 ± 3.8%), relative to the Kofun individuals (13.1 ± 3.5%) (fig. S22), as opposed to the preceding Yayoi and Kofun periods where Jomon ancestry significantly decreased due to the continental migrations (Figs. 4 and 5). Testing the genetic cladality between Kofun and Japanese by qpAdm with a “no admixture” model, we found that Kofun forms a clade with Japanese (P = 0.769). These results suggest that the genetic profile of three major ancestral components established by the state formation period has become a foundation for present-day Japanese populations, as also supported from dental and nonmetrical cranial traits (47, 48). |
|
|
|
Post by Admin on Sept 21, 2021 2:55:45 GMT
DISCUSSION Our data provide evidence of a tri-ancestry structure for present-day Japanese populations (Fig. 6), refining the established dual-structure model of admixed Jomon and Yayoi origins (5). The Jomon accumulated their own genetic variation due to long-term isolation and strong genetic drift within the Japanese archipelago following the LGM, which underlies a unique genetic component within the modern Japanese. The Yayoi period marks the end of this isolation, with substantial population migration from mainland Asia beginning at least 2.3 ka ago. However, we find a clear genetic distinction between the groups of people who arrived to the archipelago during the subsequent agrarian and state formation phases of Japanese pre- and protohistory. Genetic data from the Yayoi individuals document the presence of Northeast Asian ancestry in the archipelago as supported from morphological studies (5, 49), while we observe widespread East Asian ancestry in the Kofun. The ancestors characterizing each of the Jomon, Yayoi, and Kofun cultures made a significant contribution to the formation of Japanese populations today. 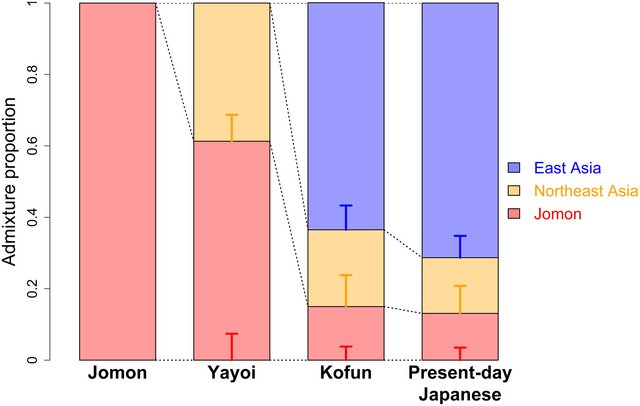 Fig. 6. Genomic transitions in parallel with cultural transitions in pre- and protohistoric Japan. The Jomon had a very unique genetic profile due to strong genetic drift and a long-term isolation in the Japanese archipelago. The rice cultivation was brought by the people who had Northeast Asian ancestry (represented by WLR_BA_o and HMMH_MN) in the Yayoi period. An additional wave of migration brought widespread East Asian ancestry (represented by Han) to the archipelago in the Kofun period. Since then, this tripartite ancestry structure has been maintained in the archipelago and become the genetic foundation of modern Japanese. The lineage ancestral to Jomon is proposed to have originated in Southeast Asia with a deep divergence from other ancient and present-day East Asians (12–14). The timing of this divergence was previously estimated to be between 18 and 38 ka ago (14); our modeling with the ROH profile of the 8.8-ka-old Jomon individual narrows this date to a lower limit within the range of 20 to 15 ka ago (Fig. 3). The Japanese archipelago had become accessible through the Korean Peninsula at the beginning of the LGM (28 ka ago) (34), enabling population movements between the continent and archipelago. The subsequent widening of the Korea Strait 17 to 16 ka ago due to rising sea levels may have led to the isolation of the Jomon lineage from the rest of the continent and also coincides with the oldest evidence of Jomon pottery production (2). Our ROH modeling also shows that the Jomon maintained a small effective population size of ~1000 during the Initial Jomon period, and we observe very little changes to their genomic profile in subsequent periods or across the different islands of the archipelago. The spread of agriculture is often marked by population replacement, as documented in the Neolithic transition throughout most of Europe, with only minimal contributions from hunter-gatherer populations observed in many regions (37–40). However, we find genetic evidence that the agricultural transition in prehistoric Japan involved the process of assimilation, rather than replacement, with almost equal genetic contributions from the indigenous Jomon and new immigrants at the Kyushu site (Fig. 4). This implies that at least some parts of the archipelago supported a Jomon population of comparable size to the agricultural immigrants at the beginning of the Yayoi period, as it is reflected in the high degree of sedentism practiced by some Jomon communities (50–53). The continental component inherited by the Yayoi is best represented in our dataset by the Middle Neolithic and Bronze Age individuals from the West Liao River basin with a high level of Amur River ancestry (i.e., WRL_BA_o and HMMH_MN) (20). Populations from this region are genetically heterogeneous in time and space (20). The Middle-to-Late Neolithic transition (i.e., between 6.5 and 3.5 ka ago) is characterized with an increase in Yellow River ancestry from 25 to 92% but a decrease in Amur River ancestry from 75 to 8% over time, which can be linked to an intensification of millet farming (20). However, the population structure changes again in the Bronze Age, which started around 3.5 ka ago, due to an apparent influx of people from the Amur River basin (fig. S17) (20). This coincides with the beginning of intensive language borrowing between Transeurasian and Sinitic linguistic subgroups (54). Excess affinity to the Yayoi is observable in the individuals who are genetically close to ancient Amur River populations or present-day Tunguisic-speaking populations (Fig. 4 and fig. S17). Our findings imply that wet rice farming was introduced to the archipelago by people who lived somewhere around the Liaodong Peninsula but who derive a major component of their ancestry from populations further north, although the spread of rice agriculture originated south of the West Liao River basin (55). The most noticeable archaeological characteristic of Kofun culture is the custom of burying the elite in keyhole-shaped mounds, the size of which reflect hierarchical rank and political power (1). The three Kofun individuals sequenced in this study were not buried in those tumuli (see note S1), which suggests that they were lower-ranking people. Their genomes document the arrival of people with majority East Asian ancestry to Japan and their admixture with the Yayoi population (Fig. 5 and fig. S17). This additional ancestry is best represented in our analysis by Han, who have multiple ancestral components. A recent study has reported that people became morphologically homogeneous in the continent from the Neolithic onward (56), which implies that migrants during the Kofun period were already highly admixed. Several lines of archaeological evidence support the introduction of new large settlements to Japan, most likely from the southern Korean peninsula, during the Yayoi-Kofun transition. Strong cultural and political affinity between Japan, Korea, and China is also observable from several imports, including Chinese mirrors and coins, Korean raw materials for iron production (1), and Chinese characters inscribed on metal implements (e.g., swords) (57). Access to these resources from overseas brought about intensive competition between communities within the archipelago; this facilitated political contact with polities in the continent, such as the Yellow Sea coast, for dominance (1). Therefore, continuous migration and continental impacts are evident throughout the Kofun period. Our findings provide strong support for the genetic exchange involved in the appearance of new social, cultural, and political traits in this state formation phase. There are caveats to this analysis. First, we are limited to only two Late Yayoi individuals from a region where skeletal remains associated with Yayoi culture are morphologically similar to Jomon (16). Yayoi individuals from other regions or other time points may have different ancestral profiles, e.g., continental-like or Kofun-like ancestry. Second, our sampling is nonrandom, as is the case of our three Kofun individuals coming from the same burial site (table S1). Additional ancient genomic data will be necessary to trace temporal and regional variation in the genetic ancestry of Yayoi and Kofun populations and provide a comprehensive view of the tripartite structure of Japanese populations proposed here. In summary, our study provides a detailed look into the changing genomic profile of the people who lived in the Japanese archipelago, both before and after agricultural and technologically driven population movements ended thousands of years of isolation from the rest of the continent. Ancient genomics on individuals from these isolated regions provides a unique opportunity to observe the magnitude of the effects of major cultural transitions on the genetic makeup of human populations. Supplementary Materials This PDF file includes: Supplementary Text Figs. S1 to S22 Tables S1 to S18 References www.science.org/doi/suppl/10.1126/sciadv.abh2419/suppl_file/sciadv.abh2419_SM.pdf |
|
|
|
Post by Admin on Sept 21, 2021 20:48:58 GMT
Late Jomon male and female genome sequences from the Funadomari site in Hokkaido, Japan
HIDEAKI KANZAWA-KIRIYAMA, TIMOTHY A. JINAM, YOSUKE KAWAI, TAKEHIRO SATO, KAZUYOSHI HOSOMICHI, ATSUSHI TAJIMA, NOBORU ADACHI, HIROFUMI MATSUMURA, KIRILL KRYUKOV, NARUYA SAITOU, KEN-ICHI SHINODA
Abstract
The Funadomari Jomon people were hunter-gatherers living on Rebun Island, Hokkaido, Japan c. 3500–3800 years ago. In this study, we determined the high-depth and low-depth nuclear genome sequences from a Funadomari Jomon female (F23) and male (F5), respectively. We genotyped the nuclear DNA of F23 and determined the human leukocyte antigen (HLA) class-I genotypes and the phenotypic traits. Moreover, a pathogenic mutation in the CPT1A gene was identified in both F23 and F5. The mutation provides metabolic advantages for consumption of a high-fat diet, and its allele frequency is more than 70% in Arctic populations, but is absent elsewhere. This variant may be related to the lifestyle of the Funadomari Jomon people, who fished and hunted land and marine animals. We observed high homozygosity by descent (HBD) in F23, but HBD tracts longer than 10 cM were very limited, suggesting that the population size of Northern Jomon populations were small. Our analysis suggested that population size of the Jomon people started to decrease c. 50000 years ago. The phylogenetic relationship among F23, modern/ancient Eurasians, and Native Americans showed a deep divergence of F23 in East Eurasia, probably before the split of the ancestor of Native Americans from East Eurasians, but after the split of 40000-year-old Tianyuan, indicating that the Northern Jomon people were genetically isolated from continental East Eurasians for a long period. Intriguingly, we found that modern Japanese as well as Ulchi, Korean, aboriginal Taiwanese, and Philippine populations were genetically closer to F23 than to Han Chinese. Moreover, the Y chromosome of F5 belonged to haplogroup D1b2b, which is rare in modern Japanese populations. These findings provided insights into the history and reconstructions of the ancient human population structures in East Eurasia, and the F23 genome data can be considered as the Jomon Reference Genome for future studies.
Introduction
The origin of modern Japanese populations has been debated among anthropologists, archaeologists, and human population geneticists. One of the main issues lacking clarity is the origin of the Jomon people. The Jomon period lasted from about 16000 to 2900 years ago (Habu, 2004; Fujio, 2015), and the Jomon people were the prehistoric natives of the Japanese islands during that period. It has long been debated whether the ancestors of the Jomon people migrated from Southeast Asia or from the northern part of East Asia.
From morphological studies of human bones and teeth, the Jomon people are thought to have originated in Southeast Asia (Turner, 1987; Hanihara, 1991; Matsumura, 2007; Matsumura and Oxenham, 2014; Matsumura et al., 2019), whereas morphological analyses (Hanihara and Ishida, 2009) and genetic studies of contemporary Japanese populations (Nei, 1995; Omoto and Saitou, 1997) have suggested that the Jomon people originated in northern Asia. However, the genetic characteristics of the Jomon people have been indirectly inferred from modern populations in Ainu and Okinawa, who were considered to have retained more Jomon ancestry than any other populations (Hanihara, 1991).
Since the 1990s, DNA retrieved from ancient human remains offer an approach that complements morphometric analysis and modern genetic data for understanding the relationships among past populations. DNA of the Jomon people was first analyzed by Horai et al. (1989). In earlier studies, mitochondrial DNA (mtDNA) was analyzed due to technical limitations of investigating low copy number DNA of ancient samples (Adachi et al., 2009). However, mtDNA is maternally inherited as a single unit without recombination. Therefore, mtDNAs are subject to chance events, such as random genetic drift, as well as gene flow and positive selection. The amount of information we can infer about social systems, such as kinship, origin, and the formation of past populations, using only mtDNA sequence data is limited.
As methods for analyzing ancient DNA have improved, whole-genome sequencing studies have become feasible (e.g. Green et al., 2010; Rasmussen et al., 2010). Next-generation sequencing technology (NGS) has enabled reconstruction of the nuclear genome of ancient human individuals. Genome-wide analysis of single nucleotide polymorphisms (SNPs) provides a powerful tool for estimating population ancestry, and many studies (e.g. Japanese Archipelago Human Population Genetics Consortium, 2012; Lazaridis et al., 2014) have shown that SNPs can be used not only to infer the geographic origin of a specific population but also to obtain information on the formation of regional groups with great accuracy. In addition, high-quality sequences of ancient nuclear genomes have enabled the inference of population size changes, social systems such as pedigree structure, risk of disease, and physical characteristics of ancient humans (e.g. Keller et al., 2012; Olalde et al., 2014; Sikora et al., 2017; Moreno-Mayar et al., 2018).
Kanzawa-Kiriyama et al. (2017) reported the first successful partial nuclear genome sequence of the Jomon people. The genome-wide analysis of ancient DNA data has provided a new source of information regarding the origins of the Jomon people. Our findings clarified that the Jomon people are phylogenetically basal to modern East Eurasians and that their DNA was inherited by modern Japanese populations. However, the depth and breadth of the nuclear genome coverage of the DNA sample analyzed in our previous work were not sufficient to fully understand the genetic characteristics of the Jomon people.
Here, we report the high-quality nuclear genome sequence of a Jomon female and a low-coverage genome sequence of a Jomon male obtained from the Funadomari site on Rebun Island, Hokkaido, Japan. The results of morphological and mtDNA analyses of the skeletal remains excavated from this site have already been reported (Matsumura et al., 2001; Adachi et al., 2009). Although Rebun Island is located at the northernmost tip of Hokkaido, the morphological characteristics (metric and nonmetric cranial traits) and dental measurements of these skeletons are similar to those of other Jomon skeletal remains from the geographically distant Honshu and Hokkaido (Matsumura et al., 2001). Additionally, the mtDNA haplogroups were also typical for Jomon individuals (Adachi et al., 2011). To obtain more detailed information about the demographic histories of the Jomon people and their relationships with contemporary human populations, we applied multiple population genetics tests using high-confidence SNPs extracted from the Funadomari Jomon DNAs. Additionally, we used functional SNP assessments to assign possible phenotypic characteristics to our sample. Analysis of Y-chromosome polymorphisms in the Funadomari male sample has also shed light on the male genealogy of the Jomon people.
|
|




