|
|
Post by Admin on Oct 2, 2021 2:31:50 GMT
Origin of Yayoi people
The PCA plot in Figure 3 clearly shows that the present-day
population genetically closest to the mainland Japanese
is Korean. Archeological evidence also supports the cultural
similarity at the beginning of the Yayoi period (~1000 BCE)
between the northern Kyusyu and the southwest Korean
peninsula (Fujio, 2017). An estimate of the split time between
present-day mainland Japanese and Koreans using
haplotype-phasing data mostly supports the hypothesis that
the split occurred about 3000 years ago (Kim et al., 2020).
However, the time estimate needs to be interpreted carefully
in general, because like all estimates based on genetic data,
it depends on assumptions of average generation time and
mutation/recombination rate in genomes.
A remaining question is how these rice-farmers migrated
to or were established in the Korean peninsula. When
rice-farming was introduced to the northern Kyushu, the
Korean peninsula was in the Mumun period, with a culture
similar to the one in the northern Kyushu (Fujio, 2017).
Studies of Y-chromosomal haplotypes revealed that the lineages of
Y-chromosomal haplogroup O, which has the highest
frequency in the mainland Japanese population, are also
frequent in Korean and Southeast Asian populations
(Hammer et al., 2006). The tie between Southeast Asia and
Japan–Korea supports the hypothesis that rice-farming originated
in the Yangtze River basin and was then introduced to
the Korean peninsula and Japanese archipelago. We tested
the hypothesis using available ancient and present-day genome data.
Ancient-genome data were obtained from the
studies of Ning et al. (2020) and Yang et al. (2020). We evaluated
whether present-day Japanese and Korean people are
genetically similar to any of the Neolithic populations linked
to the West Liao River basin, the Yellow River basin, and the
Yangtze River basin. Neolithic samples from the Yangtze
River basin were not available, and therefore we used Tanshishan
culture samples from Fujian province (Xitoucun and
Tanshishan sites, ~2500–2000 BCE) as proxy. The samples
from the West Liao River basin were BLSM45 (~3300–3100
BCE) with the Hongshan culture and EDM176 (~2000–
1600 BCE) with the Lower Xiajiadian culture. The sample
names from the Yellow River basin were WD-WT1H16
(~2100–2000 BCE) and PLTM313 (~2100–1900 BCE) with
the Henan Longshan culture, Bianbian (~7600–7500 BCE)
with the Houli culture, and Boshan (~6400–6100 BCE). The
f4 statistics showed that both present-day Japanese and Korean
populations were genetically similar to the samples from
the Early Neolithic Yellow River basin (Boshan) (Supplementary
Table 5). However, the statistical significance of the
similarity to Boshan compared with CHB was marginal
(|Z| = 2.47). On the other hand, we found Japanese–Korean
were significantly more closely related to CHB than Neolithic
southern Chinese individuals (Supplementary Table 5),
indicating that the people in the Yangtze River basin did not
substantially contribute to the genetic basis of present-day
Japanese and Korean populations. A parallel pattern to the f4
statistics is observed in the PCA plot of Figure 3; the Early
Neolithic Boshan and Bianbian individuals in the Lower
Yellow River basin were plotted fairly close to present-day
Korean individuals. Previous studies showed that the ancient
samples from North China have more northern East Asian
genetic components than the present-day Han Chinese.
The results suggest that the basal population of present-day Japanese and
Korean people was somewhat different genetically to the Late Neolithic
populations in the Yellow River and West Liao River basins. We will
know more when ancient genome data of Neolithic people in the Korean peninsula
become available.
If the dual-structure model generally holds, we would like
to know how much Jomon ancestry the present-day mainland Japanese have.
Previous studies proposed a wide range
of Jomon ancestry fractions, from 9% to 45%, using different datasets,
different models, and different assumptions
(Horai et al., 1996; Nakagome et al., 2015; Kanzawa Kiriyama et al.,
2017, 2019; McColl et al., 2018). We first
estimated the Jomon ancestry fraction using the f4 ratio test
in a similar way as Kanzawa-Kiriyama et al. (2019) did, assuming
Jomon and present-day Korean are the source populations of mainland
Japanese with Papuan and Dai as outgroup populations. With this assumption,
the estimated values were highly similar to the previous one (0.122 ± 0.009;
standard error with the block jackknife method). Because
the estimates of the admixture fraction are not robust for the
choice of outgroups, we subsequently evaluated the results
of a qpAdm analysis, using many different populations to
compute the f4 statistics (Haak et al., 2015)
|
|
|
|
Post by Admin on Oct 2, 2021 21:06:15 GMT
Yang et al. (2020) suggested the present-day Japanese are
best modeled as an admixture between the Paleolithic northern
East Asian individuals (Boshan) and Jomon individuals
(Ikawazu) with the Jomon ancestry ratio being 0.38. We
tested the model using the same source populations (Boshan
and Ikawazu) but obtained a much smaller Jomon ancestry
fraction (0.103 ± 0.030). The reason for the discrepancy is
not clear, but it is likely because we used a different set of
right (outgroup) populations to those used in the analysis by
Yang et al. (2020). When we assumed that the present-day
Japanese population was an admixture of present-day Korean and
Jomon populations (represented by F23), the estimated Jomon
ancestry by qpAdm was 0.102 ± 0.005, slightly
smaller than, but similar to, the values we obtained with the
f4 ratio test. We further tested a three-way admixture model
using the left populations (possible source populations) as
Jomon, Korean, and Hezhen, and excluded Hezhen and Oroqen from
the right populations. The three-way model (Korean, Hezhen, and Jomon, χ2
: 45.4) fits significantly better than
the two-way model (Jomon and Korean, χ2
: 58.9). The ancestry fraction of Jomon, Korean, and Hezhen was estimated to
be 0.096, 0.832, and 0.072, respectively, but the three-way
model does not fully explain the data either. If we assume that
there was no additional migration after the Yayoi period from
the Amur River basin, the pattern would imply that Yayoi
migrants might harbor more northern East Asian genetic
components than present-day Koreans do, as shown in the
previous analysis with f4 statistics. When F23 was replaced
to IK002, we obtained essentially the same results (data not
shown). In conclusion, our analysis showed that Jomon an
cestry in the present-day Japanese is around a lower bound
of the previous estimates and does not overly exceed 10%.
The 10% Jomon ancestry of present-day Japanese nuclear
genomes, however, seems to contradict the previous estimates
made using mitochondrial and Y-chromosomal genotype data.
Considering the genotype distribution of presentday Japanese and
Jomon people, mitochondrial haplogroup
M7a, N9b, and G1b, and Y-chromosome haplogroup D1a2a
(M-55) are considered to be a typical Jomon allele (Tanaka
et al., 2004; Hammer et al., 2006; Shi et al., 2008; Adachi et
al., 2009; Sato et al., 2014b). The high frequency of the
Y-chromosomal Jomon haplotype (~30%) clearly shows that
Jomon ancestry in the present-day Japanese population is
much stronger on the Y chromosomes than on autosomes
(Sato et al., 2014b). In contrast, the Jomon ancestry proportion
of mitochondrial genomes is less certain because the
frequency of M7a and N9b haplogroups in Jomon people are
somewhat variable across the Japanese archipelago (Adachi
et al., 2009). If we assumed that the proportion of M7a and
N9b haplogroups in Jomon was around 70%, the mitochondrial
Jomon ancestry would be around 15% in present-day
Japanese individuals. The observed imbalance of Jomon
ancestry among autosomal, Y-chromosomal, and mitochondrial
genomes, which we refer to as the ‘admixture paradox’,
seems confusing but worthwhile to study further to elucidate
the process of admixture of Jomon and Yayoi genetic components.
The above arguments, however, seem too simplified when
we consider a recent ancient genome sequencing study of an
approximately 2000-year-old Yayoi female from a typical
migrant culture (Shinoda et al., 2020). The analysis showed
that the genetic features of samples fell within the PCA plots
of present-day Japanese individuals, indicating that they
already had admixed genetic components of Jomon and continental
ones. The finding further complicates the story of the
origin of present-day Japanese. More ancient-genome sequencing
of populations present c. 1000 BCE in both the
Korean peninsula and Japanese archipelago could help to
explain these findings.
Conclusions
As summarized in this review, recent studies of ancientgenome
sequencing have provided us with a tremendously
important insight into the movement of prehistoric people
in Asia. Because the ancient genetic data in the Japanese
archipelago have not yet been fully analyzed, we have to
corroborate the prehistoric human movements using not
only genetic but also linguistic and archeological data
in order to reconstruct a more comprehensive history of
the region. Compared with studies in China, the genome
sequencing study of present-day and ancient people in the Japanese
archipelago is not as well developed. Even ancestry information
about present-day Japanese genome sequences has
not been thoroughly investigated yet. Future studies using
ancestry information of present-day Japanese samples and
ancient samples across prehistoric and historic time periods
would further elucidate the formation process of present-day
Japanese populations in fine detail, and could finally answer
the questions raised in Figure 1.
|
|
|
|
Post by Admin on Jan 1, 2022 20:12:14 GMT
Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations
Takashi Gakuhari, Shigeki Nakagome, …Hiroki Oota
Communications Biology volume 3, Article number: 437 (2020)
Abstract
Anatomically modern humans reached East Asia more than 40,000 years ago. However, key questions still remain unanswered with regard to the route(s) and the number of wave(s) in the dispersal into East Eurasia. Ancient genomes at the edge of the region may elucidate a more detailed picture of the peopling of East Eurasia. Here, we analyze the whole-genome sequence of a 2,500-year-old individual (IK002) from the main-island of Japan that is characterized with a typical Jomon culture. The phylogenetic analyses support multiple waves of migration, with IK002 forming a basal lineage to the East and Northeast Asian genomes examined, likely representing some of the earliest-wave migrants who went north from Southeast Asia to East Asia. Furthermore, IK002 shows strong genetic affinity with the indigenous Taiwan aborigines, which may support a coastal route of the Jomon-ancestry migration. This study highlights the power of ancient genomics to provide new insights into the complex history of human migration into East Eurasia.
Introduction
After the major Out-of-Africa dispersal of Homo sapiens around 60,000 years ago (60 kya), modern humans rapidly expanded across the vast landscapes of Eurasia1. Both fossil and ancient genomic evidence suggest that groups ancestrally related to present-day East Asians were present in eastern China by as early as 40 kya2. Two major routes for these dispersals have been proposed, either from the northern or southern parts of the Himalaya mountains1,3,4,5,6. Population genomic studies on present-day humans7,8 have exclusively supported the southern route origin of East Asian populations. On the other hand, the archaeological record provides strong support for the northern route as the origin of human activity, particularly for the arrival at the Japanese archipelago located at the east end of Eurasian continent. The oldest use of Upper Paleolithic stone tools goes back 38,000 years, and microblades, likely originated from an area around Lake Baikal in Central Siberia, are found in the northern island (i.e., Hokkaido; ~25 kya) and main-island (i.e., Honshu; ~20 kya) of the Japanese archipelago9. However, few human remains were found from the Upper Paleolithic sites in the archipelago. The Jomon culture started >16 kya, characterized by a hunter-fisher-gathering lifestyle with the earliest use of pottery in the world10. This Jomon culture lasted until a start of rice cultivation which brought by people migrated from the Eurasian continent, plausibly through the Korean peninsula, to northern parts of Kyushu island in the Japanese archipelago 3 kya. Several lines of archaeological evidence support the cultural continuity from the Upper-Paleolithic to the Jomon period, providing a hypothesis that the Jomon people are direct descendants of Upper-Paleolithic people who likely remained isolated in the archipelago until the end of Last Glacial Maximum9,11,12. Therefore, ancient genomics of the Jomon can provide new insights into the origin and migration history of East Asians.
A critical challenge for ancient genomics with samples from the Japanese archipelago is the inherent nature of warm and humid climate conditions except for the most north island, Hokkaido, and the soils indicating strong acidity because of the volcanic islands, which generally result in poor DNA preservation13,14. Though whole-genome sequences of two Hokkaido Jomon individuals dated to be 3500–3800-year-old were recently published with sufficient coverage15, a partial genome of a 3000-year-old Jomon individual from the east-north part of Honshu Japan was reported, with very limited coverage (~0.03-fold) due to the poor preservation16. To identify the origin of the Jomon people, we sequenced the genome of a 2500-year-old Jomon individual (IK002) excavated from the central part of Honshu to 1.85-fold genomic coverage. Comparing this IK002 genome with ancient Southeast Asians, we previously reported genetic affinity between IK002 and the 8000 years old Hòabìnhian hunter-gatherer17. This direct evidence on the link between the Jomon and Southeast Asians, thus, suggests the southern route origin of the Jomon lineage. Nevertheless, key questions still remain as to (1) whether the Jomon were the direct descendant of the Upper Paleolithic people who were the first migrants into the Japanese archipelago and (2) whether the Jomon, as well as present-day East Asians, retain ancestral relationships with people who took the northern route.
Here, we test the deep divergence of the Jomon lineage and the impacts of southern- versus northern-route ancestry on the genetic makeup of the Jomon. The Jomon forms a lineage basal to both ancient and present-day East Asians; this deep origin supports the hypothesis that the Jomon were direct descendants of the Upper Paleolithic people. Furthermore, the Jomon has strong genetic affinities with the indigenous Taiwan aborigines. Our study shows that the Jomon-related ancestry is one of the earliest-wave migrants who might have taken a coastal route on the way from Southeast Asia toward East Asia.
|
|
|
|
Post by Admin on Jan 1, 2022 21:37:06 GMT
The origin of IK002 To make inferences on the genetic relationship of IK002 with geographically diverse human populations, we merged the IK002 genomic data with a diverse panel of previously published ancient genomes24,25,26,27,28,29,30, as well as 300 high-coverage present-day genomes from the Simons Genome Diversity Project31. We extracted genotypes for a set of 2,043,687 SNP sites included in the “2240K” capture panel32. First, we characterized IK002 in the context of worldwide populations using principal component analysis (PCA)33,34. We found that IK002 sat in between present-day East Asians and a cluster of ancient Hòabìnhian hunter-gatherers and the Upper-Paleolithic (40 kya) individual from Tiányuán Cave17,30,35 (Fig. 1a). Second, the Honshu Jomon, IK002, closely clusters with the two Hokkaido Jomon, F23 and F5; we confirm that IK002 and the Hokkaido Jomon form a clade to the exclusion of other populations using f4-statistics (Supplementary Data 3). Henceforth, we will use IK002 as the representative of the Jomon people in this paper. Third, when using a smaller number of SNPs (41,264 SNPs) including the present-day Ainu36 from Hokkaido (Supplementary Fig. 1), IK002 clusters with the Hokkaido Ainu (Supplementary Fig. 4), supporting previous findings that the Hokkaido Ainu are direct descendants of the Jomon people16,36,37,38,39,40,41,42,43. Outgroup-f3 statistics support those PCA clustering, with IK002 sharing most genetic drift with the Hokkaido Jomon, followed by the Ainu (Supplementary Fig. 5 & Supplementary Data 4). Thus, our results indicate that IK002 is genetically distinct from present-day people in East Eurasia or even in Japan, with the exception of the Hokkaido Ainu. Fig. 1: Genetic structure of present-day and ancient Eurasian and Ikawazu Jomon. 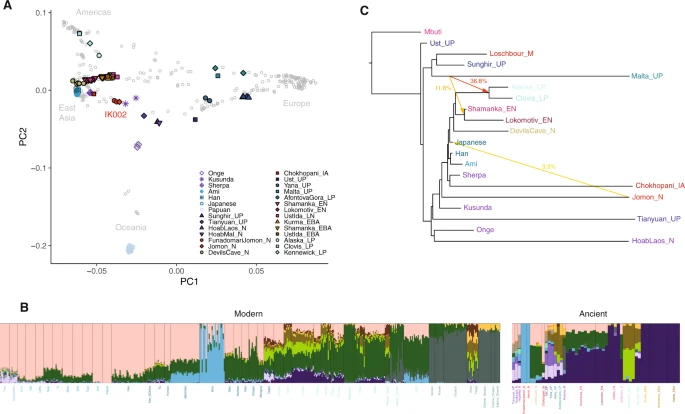 a Principal component analysis (PCA) of ancient and present-day individuals from worldwide populations after the out-of-Africa expansion. Gray labels represent population codes showing coordinates for individuals. Colored circles indicate ancient individuals. b ADMIXTURE ancestry components (K = 15) for ancient and selected contemporary individuals. The color of light blue represents the component of IK002, which is shared with the present-day Japanese and Ulchi. c Maximum-likelihood phylogenetic tree (TreeMix) with bootstrap support of 100% unless indicated otherwise. The tree shows phylogenetic relationship among present-day Southeast/East Asians, Northeast Siberians, Native Americans, and ancient East Eurasians. Mbuti are the present-day Africans; Ust’Ishim is an Upper-Paleolithic individual (45 kya) from Western Siberia83; Mal′ta (MA-1)25 and Sunghir is Upper-Paleolithic individuals (24 kya and 34 kya)29, and Loschbour is a Mesolithic individual from West Eurasia88; La368 is a pre-Neolithic Hòabìnhian hunter-gatherer (8.0 kya) in Laos and Önge is the present-day hunter-gatherers in the Andaman island, both of who are from Southeast Asia17; Tiányuán is an Upper-Paleolithic individual (40 kya) in Beijing, China35; Kusunda are the present-day minority people in Nepal; Chokhopani is an Iron-age individual (3.0–2.4 kya) and Sherpa are the present-day minority people, both of who are in Tibet6; Han, Ami and main-island Japanese are the present-day East Asians31; Devils Gate Cave is a Neolithic individual (8.0 kya) in the Primorye region of Northeast Siberia, and Shamanka and Lokomotive are Early-Neolithic individuals (8.0 kya) from Central Siberia, respectively47; USR1 and Clovis are late-Paleolithic individuals (11.5 kya and 12.6 kya) in Alaska and Montana, respectively49,89. Colored arrows represent the migration pathways and signals of admixture among all datasets. The migration weight represents the fraction of ancestry derived from the migration edge. Subsequently, we carried out model-based unsupervised clustering using ADMIXTURE44 (Supplementary Fig. 6). Assuming K = 15 ancestral clusters (Fig. 1b), an ancestral component unique to IK002 appears, which is the most prevalent in the Hokkaido Ainu (average 79.3%). This component is also shared with present-day Honshu Japanese as well as Ulchi (9.8% and 6.0%, respectively) (Fig. 1b). Those results also support the strong genetic affinity between IK002 and the Hokkaido Ainu. We used ALDER45 in order to date the timing of admixture in populations with Jomon ancestry. Using IK002 and the Hokkaido Jomon as a merged source population representing Jomon ancestry, and present-day Han Chinese as the second source representing mainland East Asian ancestry, we estimated the admixture in present-day Honshu Japanese to be between 60 and 77 generations ago (~1700–2200 years ago assuming 29 years/generation), which is slightly earlier than previous estimates8 but more consistent with the archaeological record (Supplementary Data 5). This indicates the admixture started and continuously occurred after the Yayoi period. For the Ulchi we estimated a more recent timing (31–47 generations ago) consistent with the higher variance in the IK002 component observed in ADMIXTURE. Finally, we detected more recent (17–25 generations ago) admixture for the Hokkaido Ainu, likely a consequence of still ongoing gene flow between the Hokkaido Ainu and Honshu Japanese. The estimates of admixture timing are consistent when replacing Han with Korean, Ami or Devil′s Gate cave as mainland East Asian source population, and exponential curves from a single admixture event fit the observed LD curve well (Supplementary Fig. 7 & Supplementary Data 5). To further explore the deep relationships between the Jomon and other Eurasian populations, we used TreeMix46 to reconstruct admixture graphs of IK002 and 18 ancient and present-day Eurasians and Native Americans (Fig. 1c & Supplementary Fig. 8). We found the IK002 lineage placed basal to the divergence between ancient and present-day Tibetans6,31 and to the common ancestor of the remaining ancient/present-day East Eurasians31,47,48 and Native Americans49,50. These genetic relationships are stable across different numbers of migration incorporated into the analysis. Major gene flow events recovered include the well-documented contribution of the Mal′ta individual (MA-1) to the ancestor of Native Americans25,49, as well as a contribution of IK002 to present-day mainland Japanese (m = 3–8; Supplementary Fig. 8). IK002 can be modeled as a basal lineage to East Asians, Northeast Asians/East Siberians, and Native Americans, supporting a scenario in which their ancestors arrived through the southern route and migrated from Southeast Asia toward Northeast Asia7,17. However, regarding Native Americans, high genetic contributions (11.8–36.8%) were detected from the Upper Paleolithic individual, MA-1, which means that Native Americans were admixture between the southern and the northern routes as shown in Raghavan et al. (2014). The divergence of IK002 from the ancestors of continental East Asians therefore likely predates the split between East Asians and Native Americans, which has been previously estimated at 26 kya49. Thus, our TreeMix results support the hypothesis that IK002 is a direct descendant of the people who brought the Upper Paleolithic stone tools 38,000 years ago into the Japanese archipelago. |
|
|
|
Post by Admin on Jan 1, 2022 23:59:23 GMT
The impacts of the Northern route migration into East Asia Taking advantage of the earliest divergence of the IK002 (Fig. 1c & Supplementary Fig. 8), we address a question if the Upper-Paleolithic people who took the northern route of the Himalayas mountains to arrive east Eurasia made genetic contribution to populations migrated from Southeast Asia. Under the assumption that MA-1 is a descendant of a northern route wave, we tested gene flow from MA-1 to IK002, as well as to the other ancient and present-day Southeast/East Asians and Northeast Asians/East Siberians by three different forms of D statistics: D(Mbuti, MA-1; X, Ami), D(Mbuti, X; MA-1, Ami), and D(Mbuti, Ami; X, MA-1). The first D statistics (shown as red in Fig. 2) provides results consistent with previous findings on the prevalence of MA-1 ancestry in the present-day Northeast Asians/East Siberians (Z < −3; p < 0.003, Supplementary Data 6)25, while none of Southeast/East Asians, except for Oroqen, shows a significant deviation from zero. The tree relationships observed in Fig. 1c are confirmed from the other two different forms between Ami and all of the tested populations with some variation that is mostly explained by the MA-1 gene flow (cyan and green in Fig. 2, Supplementary Data 7 & Supplementary Data 8). Therefore, there was no detectable signature of gene flow from MA-1 to the ancient/present-day Southeast/East Asians including IK002. Fig. 2: Exploring genetic affinities of IK002 within Northeast Siberians and Southeast/East Asians, respectively. 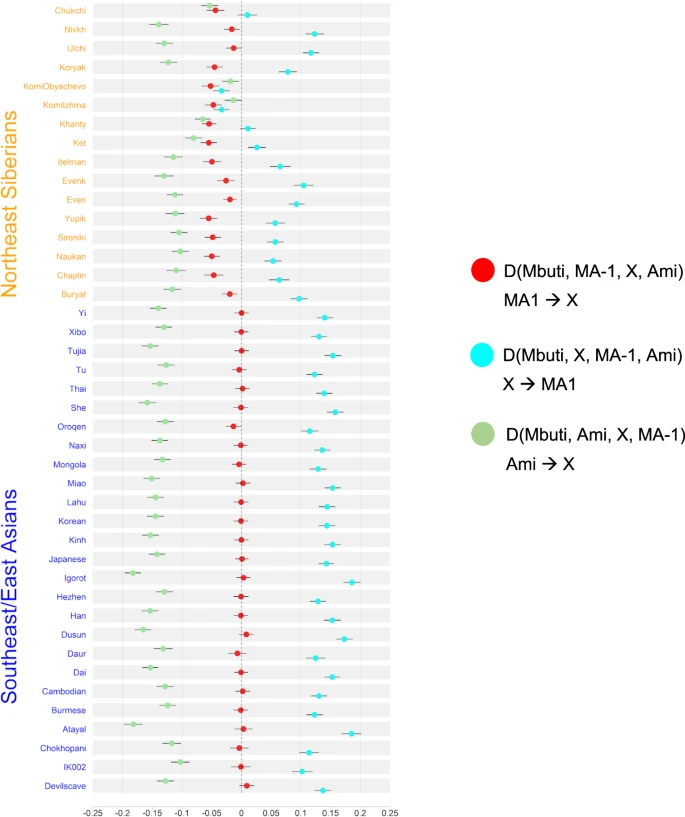 Three different D values are plotted with different colors; D(Mbuti, MA-1; X, Ami) in red, D(Mbuti, X; MA-1, Ami) in cyan, and D(Mbuti, Ami; X, MA-1) in green. Error bars show three standard deviation, and the vertical dotted and dashed lines indicate D = 0 and D = −0.2, −0.1, 0.1, and 0.2. Remnant of the Jomon-related ancestry in the coastal region The old divergence of IK002 (Fig. 1c) implies a negligible contribution to later ancient and present-day mainland East Asian groups. We further tested this implication by using f4-statistics with the form of (Mbuti, IK002; X, Chokhopani). If IK002 was a true outgroup to later East Asian groups, this statistic is expected to be zero for any test population X. However, we find that together with Japanese, present-day Taiwan aborigines (i.e., Ami and Atayal), as well as populations from the Okhotsk-Primorye region (i.e., Ulchi and Nivhk), also showed a significant (Z < −3; p < 0.003) excess of allele sharing with IK002. Populations in the inland of the eastern part of the Eurasian continent on the other hand were consistent with forming a clade with Chokhopani (Fig. 3), which suggests the presence of Ikawazu Jomon-related ancestry in the present-day coastal populations in East Asia. The signal is also present in the Neolithic individuals from Devil′s Gate Cave in the Primorye region (Z < −3; p < 0.003; Supplementary Fig. 9), suggesting that, at 8 kya, populations who had IK002-related ancestry in the region had already been largely but not completely replaced by later migrations. Interestingly, the genetic affinity to IK002 was found only in the coastal region but not in the inland for both ancient and present-day populations (Fig. 3). Fig. 3: Heatmap of f4-statistics comparing Eurasian populations to the Ikawazu Individual. 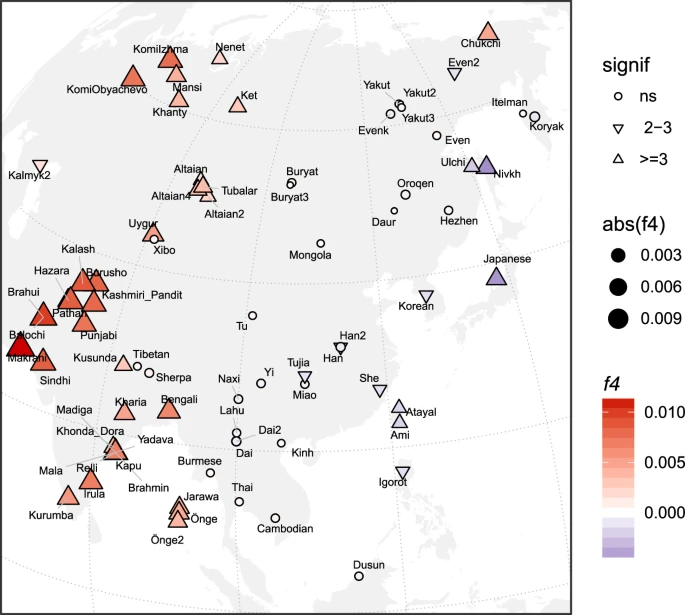 Heatmaps of f4(Mbuti, IK002;X, Chokhopani), where X are the present-day East Eurasian populations. The color and size represents the value of f4-statistics. The shape represents statistical significances of genetic affinities based on Z-score. Triangle label means statistical significance with |Z| > 3 (P < 0.01), inverted triangle means weak significance with |Z| = 2–3 and circle means non-significance with |Z| < 2 (P > 0.05). We carried out admixture graph modeling to further characterize the contributions of IK002-related ancestry to other East Asian populations. To that end, we first fit a backbone graph including ancient genomes representative of major divergences among East Asian lineages: IK002 (early dispersal); Chokhopani (later dispersal, East Asia), and Shamanka (later dispersal, Siberia). Test populations of interest were then modeled as three-way mixtures of early (IK002) and later (Chokhopani, Shamanka) dispersal lineages, using a grid search of admixture proportions within qpGraph. Consistent with the results from the f4-statistics, we find that models without contribution from IK002 result in poor fit scores for Japanese, Devil′s Gate Cave and Ami, as opposed to inland groups such as Han which do not require IK002-related ancestry (Supplementary Fig. 10). The range of admixture fractions with good model fit is generally quite wide, with best fit models showing IK002-related contributions of 8%, 4 and 41% into Japanese, Devil′s Gate Cave and Ami, respectively (Supplementary Fig. 10 & Supplementary Fig. 11). While the substantial contribution into Ami seems at odds with the lower f4-statistics compared with Japanese, the lineage admixing with Ami shares only a very short branch with IK002, suggesting a contribution from a distinct group with an early divergence from the IK002 lineage. We note that this backbone graph fitting assumed an unadmixed Jomon lineage, as opposed to a previously suggested dual-ancestry model where Jomon is admixed between Önge- and Ami-related ancestry. This alternative base model provides an equivalent admixture graph fit, however we find no evidence for shared genetic drift between the Önge and the ancestral Jomon lineage in qpGraph (Supplementary Fig. 12a & Supplementary Fig. 12b), or using direct f4-statistics (Supplementary Data 9 & Supplementary Data 10). Additional sampling of early East Asian human remains will be needed to further resolve the relationships among these deep lineages, but nevertheless either model supports the deep origins of Jomon. |
|



