Post by Admin on Aug 25, 2018 19:44:33 GMT
Discussion
This is the first study on the ancient nuclear genome of the Neolithic hunter-gatherers who lived on the Japanese Archipelago. We observed more post-mortem changes in the Sanganji Jomon sample than in two other archaic human examples (~0.8% in Sanganji Jomon, Supplementary Table S7). Environmental conditions (for example, temperature, humidity, pH value, geochemical properties of soil) or use of different sample treatments may affect the condition of the ancient DNA. We did not treat the ancient DNA with uracil DNA glycosylase and endonuclease VIII, which remove C to T misincorporation from the sequence reads. This is to reduce the number of experimental steps so as to minimize the risk of contamination in library preparations. We also chose to see the post-mortem damage specific to ancient DNA to more clearly evaluate the ancient origin of the extracted DNA. This may be the reason that the DNA damage was high in Sanganji Jomon sequences.
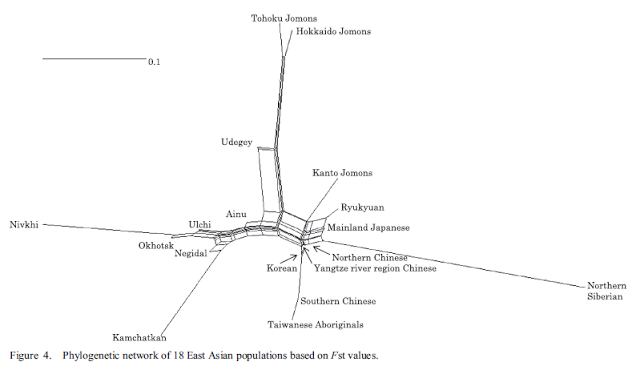
PCA and phylogenetic analyses revealed the genetic relationships between the Sanganji Jomon and the modern human populations that we compared. We found that Sanganji Jomon was genetically quite distinct from the other modern East Eurasians. Our Jomon genotypes are still mostly based on a single read per SNP site, whereas the 1000 Genomes Project data24 had at least 4–6 × coverage. We need to be careful about the effect of genotype error rate even if we use only transversion sites (error rate=0.0129%). However, PCA plots using all sites (Figure 1 and Supplementary Figures S7 and S9) and those using transversion only (Supplementary Figures S8) are more or less the same, and TreeMix trees using all sites (Figure 4 and Supplementary Figures S16 and S18) and those using transversion only (Supplementary Figure S17) also showed similar positions for the Sanganji Jomon. Therefore, we believe that the phylogenetic relationship of the Sanganji Jomon can still be reliably estimated with our data.
Some cranial metric and dental analyses suggested that the ancestral population of the Jomon people originated from Southeast Asia,3, 39, 40, 41, 42, 43 whereas other morphological analyses44, 45 and mtDNA sequence data9 suggested that their ancestors were of Northeast Asian origin. However, neither hypothesis was supported from our analysis using direct Jomon nuclear genome sequences. Our results suggest that the Jomon people were descendants of an ancestral East Eurasian population prior to population diversifications recognizable today.
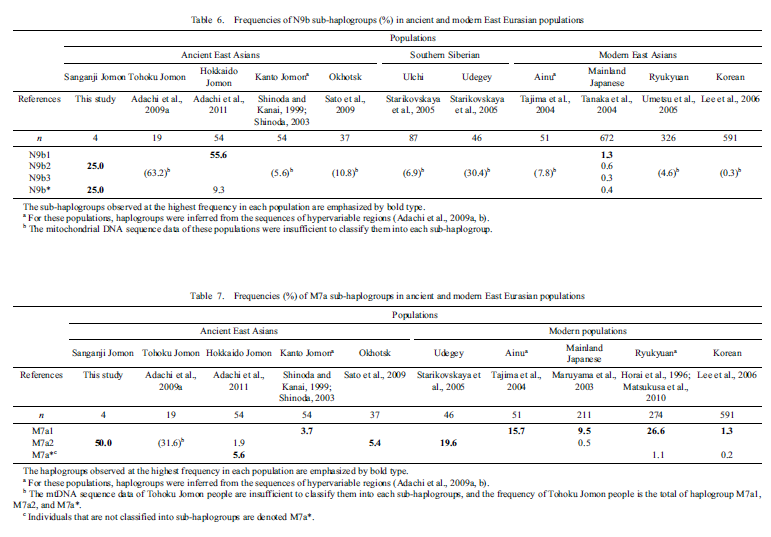
The possibility that the Sanganji Jomon ancestors diverged before the Native Americans was also inferred through the maximum-likelihood tree, neighbor-joining tree, and the D-statistic tests, but this is still inconclusive because of the small number of SNPs in the analyses. However, all the results suggest a deep Jomon divergence within East Asia, close to the Native American split. The effect of genotype errors also need to be taken into account in interpreting the apparent deep divergence of the Jomon people. A relatively large amount of genotype errors in the Jomon data set would induce their long tree branch and/or genetic similarity between the Jomon and outgroups (for example, non-East Eurasians and archaic humans) as artifacts, placing the Jomon closer to the root of outgroup-East Eurasian splits and/or showing gene flow between Jomon and those outgroups. However, since we did not observe any of those artifacts, and the inferred genotype error rate in the transversion-only sites (0.0129%) is quite small, it is unlikely that the Jomon was erroneously located at the basal position of East Eurasians. The ages of the most recent common ancestors of mitochondrial DNA haplogroups N9b and M7a (considered indigenous to Jomon and modern Japanese, and called the Jomon haplotype), were estimated to be 22 000 YBP and 23 000 YBP, respectively.10 These estimates also support a deep divergence of the Jomon people.
We also examined whether the uniqueness of the Sanganji Jomon in East Eurasia was the result of gene flow from non-East Eurasians, but the evidence for this was not suggested (Supplementary Figures S22, S24 and S25). This indicates that the ancestor of the Sanganji Jomon had been genetically isolated from continental populations after their divergence. However, our results do not deny a possibility of small fraction of gene flow between the Jomon people and the non-East Eurasians, which might not be detected in the current study because of the small amount of the determined Jomon genome sequences. The TreeMix tree and the D-statistic tests also confirm the lack of non-East Eurasian contaminations in the Jomon data via reagents because if there were such contamination during experiments, the effect would be artificially detected as gene flow in those analyses. Genetic affinity between modern East Eurasians and Melanesian compared with Sanganji Jomon was inferred, but these results were also ambiguous because of the data limitation. Further genomic sequencing of Jomon genomes in future studies will clarify such questions.
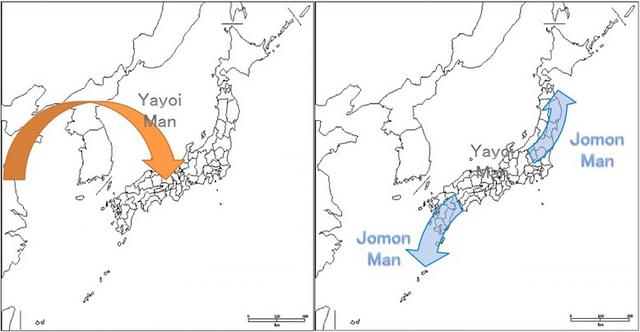
Our results show that the populations genetically closest to Sanganji Jomon are those that live on the Japanese Archipelago. The current Japanese population is widely accepted to be a result of admixture of indigenous Jomon and later migrant people, whereby the latter were agricultural people that came from continental Asia into the Japanese Archipelago probably via the Korean Peninsula during and after the Yayoi era. Until now, this hybridization scenario has not been verified directly at the genomic level. Our result based on PCA, TreeMix and neighbor-joining trees, Neighbor-Net networks, and D-statistic tests clarified that the Jomon people genetically contributed to the modern Japanese Archipelago populations. The genetic similarity would not be explained by contamination from Japanese archipelago populations during experiments, because the direction of gene flow estimated in TreeMix tree was from Jomon to JPT, not from JPT to Jomon (Figure 4), and the D((KHV or CHS, CHB) Jomon) value in D-statistic tests (Figure 6a) was not negative with significant Z-score (JPT was genetically closer to CHB than to CHS or KHV). Furthermore, if there was contamination from modern Japanese, the Jomon would be genetically closer to CHB than to CHS and KHV, leading to negative D-values and probably significant Z-scores. Therefore, we can conclude that the modern Japanese Archipelago populations are the admixture of the Jomon people and other populations with genetic affinities with modern Northeast Asians. This result is consistent with the conclusion based on the genetic analyses of modern human populations.1 However, it is difficult to pin-point the geographic origins of the candidate populations because of lack of genetic data of relevant ancient populations.
The amount of genetic contribution from the Jomon people varied among the Japanese Archipelago populations (Figures 2b and 3a and Supplementary Figure S15). The Ainu and the Ryukyuan share more alleles with the Jomon than the mainland Japanese, suggesting smaller genetic contributions from continental populations in those two populations than for the mainland Japanese, in agreement with previous findings.1, 2, 4, 13, 46, 47, 48, 49, 50 This supports the dual-structure model of Hanihara,3, 50 which is a widely accepted theory that explains the history of the three current populations that inhabit the Japanese Archipelago, and which predicts that the mainland Japanese are more admixed with agricultural continental people than the Ainu and the Ryukyuan, with major admixture occurring in and after the Yayoi period.
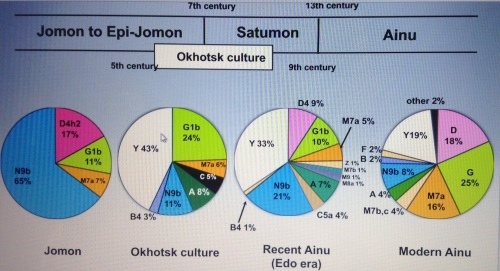
PC1 values of some Ainu individuals were similar to that of Sanganji Jomon, whereas PC2 separated the Ainu people from Sanganji Jomon (Figure 2b). It is probable that the ancestors of the Ainu people experienced admixture with other population(s) (for example, continental population(s) not used in the current study) after the Jomon period, and they are thus genetically differentiated from the Jomon people.51 The candidate population is the Okhotsk people, who inhabited the Northeastern costal area of the Hokkaido region, Japan, from the 5th to 13th century. They were morphologically close to modern southern Siberians such as the Nivkhi and Ulchi people,52, 53, 54, 55 who were not included in the current study. Recent studies suggest that the descendants of the Hokkaido Jomon people had admixed with the Okhotsk people and became ancestors of the modern Ainu.56, 57, 58, 59
Jinam et al.2 recently estimated the proportion of the Jomon ancestry in the mainland Japanese using the f4-ratio test35under a specified phylogenetic scenario. The estimated proportion of Jomon ancestry differs depending on the compared populations and were within the range of 13–21%. Jinam et al.2 also used ADMIXTURE60 and found that the mainland Japanese was best explained by two ancestry components. One of them has very high proportions in many Ainu individuals, and is considered to be Jomon ancestry if we assume that the Ainu people are largely descendants of the Jomon. The average proportion of this Jomon component in the mainland Japanese was 17%, which is within the range of f4-ratio test results.2 The same genome-wide SNP data were also used for ABC analysis, and the highest likelihood model estimated the admixture proportion from the Jomon ancestry to the mainland Japanese to be 36% (22–53%, 95% confidence interval).50 We used TreeMix for estimating Jomon ancestry proportions in this study, and the frequency was 12% as shown in Figure 4. We note that it is possible that the frequency was underestimated if the Jomon people had some population substructure and the northern Jomon including the Sanganji Jomon only indirectly contributed to the modern Japanese, and/or if there are contamination during experiments from the Japanese Archipelago populations. Previously, the proportion of the Jomon component in the modern mainland Japanese was estimated to be 35% based on partial mtDNA sequences of modern human populations46and 23–40% based on application of a demographic model of population admixture.61 Although the Jomon admixture proportion is still debatable, we would like to conclude that the Jomon component in the mainland Japanese is probably lower than 20%.
Journal of Human Genetics volume62, pages213–221 (2017)
This is the first study on the ancient nuclear genome of the Neolithic hunter-gatherers who lived on the Japanese Archipelago. We observed more post-mortem changes in the Sanganji Jomon sample than in two other archaic human examples (~0.8% in Sanganji Jomon, Supplementary Table S7). Environmental conditions (for example, temperature, humidity, pH value, geochemical properties of soil) or use of different sample treatments may affect the condition of the ancient DNA. We did not treat the ancient DNA with uracil DNA glycosylase and endonuclease VIII, which remove C to T misincorporation from the sequence reads. This is to reduce the number of experimental steps so as to minimize the risk of contamination in library preparations. We also chose to see the post-mortem damage specific to ancient DNA to more clearly evaluate the ancient origin of the extracted DNA. This may be the reason that the DNA damage was high in Sanganji Jomon sequences.
PCA and phylogenetic analyses revealed the genetic relationships between the Sanganji Jomon and the modern human populations that we compared. We found that Sanganji Jomon was genetically quite distinct from the other modern East Eurasians. Our Jomon genotypes are still mostly based on a single read per SNP site, whereas the 1000 Genomes Project data24 had at least 4–6 × coverage. We need to be careful about the effect of genotype error rate even if we use only transversion sites (error rate=0.0129%). However, PCA plots using all sites (Figure 1 and Supplementary Figures S7 and S9) and those using transversion only (Supplementary Figures S8) are more or less the same, and TreeMix trees using all sites (Figure 4 and Supplementary Figures S16 and S18) and those using transversion only (Supplementary Figure S17) also showed similar positions for the Sanganji Jomon. Therefore, we believe that the phylogenetic relationship of the Sanganji Jomon can still be reliably estimated with our data.

Some cranial metric and dental analyses suggested that the ancestral population of the Jomon people originated from Southeast Asia,3, 39, 40, 41, 42, 43 whereas other morphological analyses44, 45 and mtDNA sequence data9 suggested that their ancestors were of Northeast Asian origin. However, neither hypothesis was supported from our analysis using direct Jomon nuclear genome sequences. Our results suggest that the Jomon people were descendants of an ancestral East Eurasian population prior to population diversifications recognizable today.
The possibility that the Sanganji Jomon ancestors diverged before the Native Americans was also inferred through the maximum-likelihood tree, neighbor-joining tree, and the D-statistic tests, but this is still inconclusive because of the small number of SNPs in the analyses. However, all the results suggest a deep Jomon divergence within East Asia, close to the Native American split. The effect of genotype errors also need to be taken into account in interpreting the apparent deep divergence of the Jomon people. A relatively large amount of genotype errors in the Jomon data set would induce their long tree branch and/or genetic similarity between the Jomon and outgroups (for example, non-East Eurasians and archaic humans) as artifacts, placing the Jomon closer to the root of outgroup-East Eurasian splits and/or showing gene flow between Jomon and those outgroups. However, since we did not observe any of those artifacts, and the inferred genotype error rate in the transversion-only sites (0.0129%) is quite small, it is unlikely that the Jomon was erroneously located at the basal position of East Eurasians. The ages of the most recent common ancestors of mitochondrial DNA haplogroups N9b and M7a (considered indigenous to Jomon and modern Japanese, and called the Jomon haplotype), were estimated to be 22 000 YBP and 23 000 YBP, respectively.10 These estimates also support a deep divergence of the Jomon people.
We also examined whether the uniqueness of the Sanganji Jomon in East Eurasia was the result of gene flow from non-East Eurasians, but the evidence for this was not suggested (Supplementary Figures S22, S24 and S25). This indicates that the ancestor of the Sanganji Jomon had been genetically isolated from continental populations after their divergence. However, our results do not deny a possibility of small fraction of gene flow between the Jomon people and the non-East Eurasians, which might not be detected in the current study because of the small amount of the determined Jomon genome sequences. The TreeMix tree and the D-statistic tests also confirm the lack of non-East Eurasian contaminations in the Jomon data via reagents because if there were such contamination during experiments, the effect would be artificially detected as gene flow in those analyses. Genetic affinity between modern East Eurasians and Melanesian compared with Sanganji Jomon was inferred, but these results were also ambiguous because of the data limitation. Further genomic sequencing of Jomon genomes in future studies will clarify such questions.
Our results show that the populations genetically closest to Sanganji Jomon are those that live on the Japanese Archipelago. The current Japanese population is widely accepted to be a result of admixture of indigenous Jomon and later migrant people, whereby the latter were agricultural people that came from continental Asia into the Japanese Archipelago probably via the Korean Peninsula during and after the Yayoi era. Until now, this hybridization scenario has not been verified directly at the genomic level. Our result based on PCA, TreeMix and neighbor-joining trees, Neighbor-Net networks, and D-statistic tests clarified that the Jomon people genetically contributed to the modern Japanese Archipelago populations. The genetic similarity would not be explained by contamination from Japanese archipelago populations during experiments, because the direction of gene flow estimated in TreeMix tree was from Jomon to JPT, not from JPT to Jomon (Figure 4), and the D((KHV or CHS, CHB) Jomon) value in D-statistic tests (Figure 6a) was not negative with significant Z-score (JPT was genetically closer to CHB than to CHS or KHV). Furthermore, if there was contamination from modern Japanese, the Jomon would be genetically closer to CHB than to CHS and KHV, leading to negative D-values and probably significant Z-scores. Therefore, we can conclude that the modern Japanese Archipelago populations are the admixture of the Jomon people and other populations with genetic affinities with modern Northeast Asians. This result is consistent with the conclusion based on the genetic analyses of modern human populations.1 However, it is difficult to pin-point the geographic origins of the candidate populations because of lack of genetic data of relevant ancient populations.

The amount of genetic contribution from the Jomon people varied among the Japanese Archipelago populations (Figures 2b and 3a and Supplementary Figure S15). The Ainu and the Ryukyuan share more alleles with the Jomon than the mainland Japanese, suggesting smaller genetic contributions from continental populations in those two populations than for the mainland Japanese, in agreement with previous findings.1, 2, 4, 13, 46, 47, 48, 49, 50 This supports the dual-structure model of Hanihara,3, 50 which is a widely accepted theory that explains the history of the three current populations that inhabit the Japanese Archipelago, and which predicts that the mainland Japanese are more admixed with agricultural continental people than the Ainu and the Ryukyuan, with major admixture occurring in and after the Yayoi period.
PC1 values of some Ainu individuals were similar to that of Sanganji Jomon, whereas PC2 separated the Ainu people from Sanganji Jomon (Figure 2b). It is probable that the ancestors of the Ainu people experienced admixture with other population(s) (for example, continental population(s) not used in the current study) after the Jomon period, and they are thus genetically differentiated from the Jomon people.51 The candidate population is the Okhotsk people, who inhabited the Northeastern costal area of the Hokkaido region, Japan, from the 5th to 13th century. They were morphologically close to modern southern Siberians such as the Nivkhi and Ulchi people,52, 53, 54, 55 who were not included in the current study. Recent studies suggest that the descendants of the Hokkaido Jomon people had admixed with the Okhotsk people and became ancestors of the modern Ainu.56, 57, 58, 59
Jinam et al.2 recently estimated the proportion of the Jomon ancestry in the mainland Japanese using the f4-ratio test35under a specified phylogenetic scenario. The estimated proportion of Jomon ancestry differs depending on the compared populations and were within the range of 13–21%. Jinam et al.2 also used ADMIXTURE60 and found that the mainland Japanese was best explained by two ancestry components. One of them has very high proportions in many Ainu individuals, and is considered to be Jomon ancestry if we assume that the Ainu people are largely descendants of the Jomon. The average proportion of this Jomon component in the mainland Japanese was 17%, which is within the range of f4-ratio test results.2 The same genome-wide SNP data were also used for ABC analysis, and the highest likelihood model estimated the admixture proportion from the Jomon ancestry to the mainland Japanese to be 36% (22–53%, 95% confidence interval).50 We used TreeMix for estimating Jomon ancestry proportions in this study, and the frequency was 12% as shown in Figure 4. We note that it is possible that the frequency was underestimated if the Jomon people had some population substructure and the northern Jomon including the Sanganji Jomon only indirectly contributed to the modern Japanese, and/or if there are contamination during experiments from the Japanese Archipelago populations. Previously, the proportion of the Jomon component in the modern mainland Japanese was estimated to be 35% based on partial mtDNA sequences of modern human populations46and 23–40% based on application of a demographic model of population admixture.61 Although the Jomon admixture proportion is still debatable, we would like to conclude that the Jomon component in the mainland Japanese is probably lower than 20%.
Journal of Human Genetics volume62, pages213–221 (2017)