Post by Admin on Mar 31, 2019 17:43:32 GMT
Links to ancient genomes
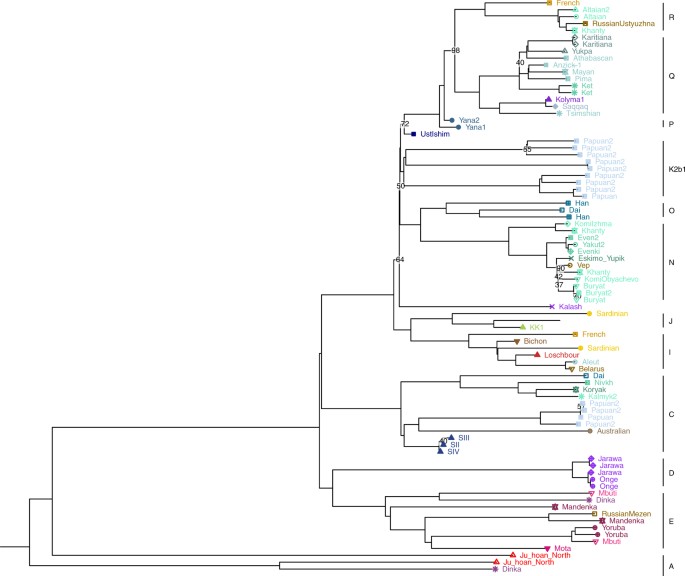
Genetic affinity between modern Siberians and Ust'-Ishim
To uncover ancient demographic events and reveal genetic links between present-day individuals and ancient ancestral populations, we compared 60 genomes from modern-day humans (Table 1; Supplemental Table S2) to 46 ancient genomes (Supplemental Table S3), including a 45,000-yr-old Siberian Ust’-Ishim individual (Fu et al. 2014), a 24,000-yr-old Siberian Mal'ta boy (MA-1), and a 17,000-yr-old Siberian individual (AG-2) from the Krasnoyarsk region (Raghavan et al. 2014).
Given that the bone from the Ust’-Ishim individual was found in Western Siberia, we reasoned that Ust’-Ishim might be related to modern Siberian populations. Indeed, this hypothesis was well supported by the TreeMix model (Fig. 6). We observed that the common ancestor of Siberians and East Asians traced 38% (95% CI: 28%–48%) of their ancestry to Ust’-Ishim's lineage (Fig. 6; Supplemental Fig. S14). In agreement with this, the D-statistics analysis (Fig. 5C; Supplemental Figs. S13C, S15) showed that Ust’-Ishim had higher genetic affinity with East Asians and Siberians, particularly Eastern Siberians, than with modern Europeans. This was also supported by the Y haplotype analysis. Previously, the ancient Siberian Ust’-Ishim (Fu et al. 2014) was thought to belong to the K2 Y-Chromosome clade, which is ancestral to haplogroup R (a Y-Chromosome clade extremely common in Europe), haplogroup Q (common among Native Americans and certain Siberians populations), haplogroup N (common in Siberia and Northeastern Europe), and haplogroup O (common in East Asia). However, we observed that Ust’-Ishim had a single derived SNP on Y-Chromosome specific to the NO branch (hg19 coordinate Chr Y 7,690,182) (Supplemental Fig. S12). This branch is associated with Siberians and some Northeastern European groups, as well as East Asians. Therefore, unlike previous claims, Ust’-Ishim's haplogroup most likely belongs to a more recent NO clade rather than to the more diverged K2 clade. Based on these lines of evidence, we conclude that Ust’-Ishim contributed more ancestry to the East Asian and Siberian populations than to modern Europeans or Native Americans.
Western Siberians are descendants of ANEs and Eastern Siberians
Our tree model grouped ANEs MA-1 and AG-2 with Western Siberians (Mansi and Nenets), suggesting a genetic link between ANEs and modern Western Siberians (Fig. 7). Furthermore, TreeMix estimated that 41% (95% CI: 36%–45%) of the ancestry of Andean Highlanders, the Native Americans, is attributable to the ANE lineage (Fig. 7; Supplemental Fig. S16). This is consistent with previous reports that Paleo-Americans trace ∼42% of their autosomal genome to an ancient Eurasian lineage related to MA-1 (Raghavan et al. 2014). We further tested these predictions using D-statistics by evaluating whether a modern individual shared significantly more derived alleles with MA-1 or AG-2 than with the East Asian Han individual (Fig. 5D; Supplemental Fig. S17A). Mansi, Khanty, Nenets, and Native Americans all had very strong ANE ancestry, demonstrated by their significant genetic affinities with MA-1 and AG-2. Therefore, Western Siberians share common ANE ancestry with Native Americans. The remaining ancestry of Western Siberians could be attributed to a population related to Eastern Siberians. The TreeMix model (Fig. 7) inferred that 43% of Mansi ancestry was derived from the admixture with a population related to Eastern Siberians Evenks and Evens, while 57% is attributable to the ANE ancestry. Therefore, genomes of Mansi people harbor significant ancestries from Siberian ANE people as well as Eastern Siberian populations in approximately equal proportions.

Figure 7.
Genetic relationships with ancient North Eurasians MA-1 and AG-2. Autosomal TreeMix admixture graph that includes ancient 7000-yr old La Brana (Spain) and low-coverage genomes of ancient Siberians MA-1 and AG-2. Residual plot for this graph is shown in Supplemental Figure S16. TreeMix model indicated multiple admixture events: 41% admixture (95% CI: 36%–45%) from the ANE clade represented by MA-1 and AG-2 individuals into Native Americans represented by Andean Highlanders, 43% admixture (95% CI: 38%–47%) between population related to Evenk and a common ancestor of Western Siberians (Mansi and Nenets), and 26% (95% CI: 16%–37%) admixture between population related to Even and modern-day Nenets. Additionally, Western Siberians and ANE are grouped together, indicating their shared genetic history.
To examine Siberian-related ancestry among ancient Europeans, we analyzed SNP data from the Holocene Eastern European hunter-gatherers (Haak et al. 2015) from the Samara and Karelia regions in modern Russia and the Motala region in Sweden (6.6–8 kya), as well as more recent Bronze Age Yamnaya samples (4.7–5.3 kya). D-Statistics demonstrated strong admixtures between the Mansi and nearly all hunter-gatherers from Eastern Europe (Supplemental Fig. S21), particularly for the Samara and Karelia samples. Slightly weaker, but significant affinities were present between Eastern Siberian Even and Eastern European hunter-gatherers. As previously mentioned, the time of the split of Eastern and Western Siberian populations was estimated to be around 6.8–9.9 kya. The weaker affinity of Eastern Siberians and Eastern European hunter-gatherers could be attributed to the Eastern Siberian ancestry of Mansi, which permeated into ancient European populations through Mansi-related admixture. Yamnaya samples also showed statistically significant admixtures with Mansi and Evens, but slightly weaker compared with Samara HG, which indicates a dilution of Eastern hunter-gatherer ancestry component of Yamnaya culture samples. The ANE-related ancestry among Eastern European hunter-gatherers could thus be attributed to gene flows between population ancestral to Mansi and Eastern European hunter-gatherers that occurred before 8 kya, as demonstrated by the strong genetic affinity between Mansi and Samara and Karelia hunter-gatherers from 6–8 kya.
Apart from Eastern European hunter-gatherers, Siberians also shared part of their ancestry with Pitted Ware Culture (PWC) 5,000-yr-old hunter-gatherers from Sweden Ire8 and Ajv52 (Skoglund et al. 2012). Ajv52 and particularly Ire8 had strong admixture signals with Western Siberians Mansi (Fig. 5E; Supplemental Fig. S17B), suggesting that like the closely related Yamnaya culture, they had strong ANE ancestries likely due to admixtures with Mansi-related population.
Shared ancestry between Eastern Siberians and ancient human of the Saqqaq culture
Our data also showed evidence of Eastern Siberian ancestry within the genome of a 4000-yr-old Saqqaq individual (Rasmussen et al. 2010) from Greenland. The tree model with Saqqaq genome showed that Saqqaq was related to both East Asians and Eastern Siberians, and 12% of its ancestry was attributable to a population related to modern-day Evens (Supplemental Figs. S19, S20). The detected gene flow was strongly supported by the D-statistics (Supplemental Figs. S7F, S8F). In addition, 18% of American Andeans' ancestry came from Evens (Supplemental Fig. S19). This, together with the substantial gene flow from an ancestor of Evens and Evenks to Altayans, suggests that the signal from Evens to Saqqaq can be a result of ancient admixture happened between the ancestor of Siberians (Eastern Siberians) and early migrants to the Americas.

Figure 8.
Geographical dispersion model. The approximate model of gene flows reflecting divergence of primary clades in Europe, East Asia, and Siberia with approximate divergence times based on Chromosome Y, MSMC, TreeMix, and D-statistics analyses. Percentages represent proportions of ancestry contributed to each lineage from other lineages. The common ancestors of Mansi, Khanty, and Nenets had derived 57% of their ancestry from an ANE population, related to MA-1, AG-2, and Native Americans and 43% from the admixture with a population related to Evens and Evenks. Native Americans trace 42% of their ancestry to ANE and 58% to a common ancestor of Eastern Siberians and East Asians. In the figure, 45 kya represents the split between Y-Chromosome clades QR and NO; 33 kya represents the split between Y-Chromosome Q and R clades and mtDNA N2 and W clades; 17 kya represents Mansi-Andean separation time; 44 kya represents the split of Ust’-Ishim's haplogroup from NO clade; 19 kya represents Han-Andean separation time; 10 kya represents Han-Eastern Siberian separation time; and 5.3–9.9 kya represents Evenk-Mansi MSMC separation time (6.8–9.9 kya) and expansion of N1c1 Y-haplogroup (5.3–7.1 kya). In this figure, Europeans represent a collective term for the following populations from this study: Komi, Veps, Karelians, Russians, and Belarussians.
Discussion
Genomes of indigenous Siberians harbor evidence of many important ancient events that shaped population history and peopling of Eurasia and the New World. Siberia is home to more than 30 ethnic groups that were thus far undersurveyed in population genetic studies. In this work, we examined the genetic history of several Siberian and Eastern European populations and analyzed their genetic links to ancient and modern populations. We summarized our findings in the form of a synthetic geographical dispersion model (Fig. 8), which describes how populations and genetic variation likely spread across Northern Eurasia.
Our analyses demonstrated that both East Asian and Siberian populations shared a significant amount of ancestry (38%) attributable to the ancient Siberian Ust’-Ishim individual, thus pointing to very ancient origins of indigenous Siberians. D-Statistics tests also suggested stronger affinity of Ust’-Ishim to both Eastern and Western Siberians (Fig. 5C; Supplemental Figs. S13C, S15) than to modern Europeans. Another line of evidence pointing to affinity of both East Asians and Eastern Siberians with Ust’-Ishim was provided by the Y-Chromosome haplogroup analysis (Fig. 4A; Supplemental Fig. S12). We showed that Ust’-Ishim's Y-Chromosome haplogroup belonged to NO clade, and its divergence from NO lineage predated the split of Y-Chromosome haplogroup N (particularly common in Siberia and Northeastern Europe) and haplogroup O (the most common Y-Chromosome haplogroup among Eastern and Southern Asian populations). These new relationships were uncovered largely due to the inclusion of the Western and Eastern Siberian genomes, including Mansi, Khanty, Nenets, Evens, Buryat, and other Siberian individuals. Therefore, the new findings described here provide a more accurate interpretation of patterns observed in the original Ust’-Ishim study (Fu et al. 2014).
A particularly surprising finding was the genetic link between 24,000-yr-old Siberian Mal'ta boy, Native Americans, and the Western Siberians Mansi, Khanty, and Nenets. This finding demonstrates a strong genetic link between Western Siberians and Native Americans due to their common ANE ancestry. We estimate that 57% of Western Siberian Mansi and Khanty ancestry was related to the ANE lineage represented by MA-1 and AG-2 ancient individuals. The other important finding was that the remaining 43% of Western Siberian ancestry was related to Eastern Siberian populations, suggesting admixtures between Eastern Siberian groups most related to Evens and Evenks and the common ancestors of Western Siberians (Fig. 2). This is particularly interesting in the context of ancestral relationships of modern Northeastern European populations. Lazaridis et al. (2014) modeled the ancestry of Eastern Europeans such as Mordovians, Finns, Russians, Saami, and Chuvashs but could not completely explain their genetic makeup by a mixture of three early European groups: the ancient northern Eurasians (represented by MA-1), the West European hunter-gatherers (represented by Loschbour), and the early European farmers (represented by Stuttgart). This was proposed to be due to a greater relatedness of Eastern Europeans to East Asians, compared with other European groups. However, our analyses of genetic relationships of Northeastern European populations such as Mezen Russians, Komi, Karelians, and Veps suggested that the unexplained ancestral components among modern Europeans likely resulted from their affinity with both Western and Eastern Siberian populations.
Our findings also showed that Western Siberians Mansi and Khanty and ancient Eastern European populations (Yamnaya and the PWC people) shared a significant amount of ANE-related ancestry. The genetic affinity of ancient Eastern Europeans with Eastern Siberians is relatively weaker (Fig. 5E; Supplemental Figs. S21, S24). Therefore, we propose that the Western Siberian admixture into Northeastern Europeans likely began before the Yamnaya culture period (5.3–4.7 kya), since the admixtures with Mansi are also very strong among hunter-gatherers from Northeastern Europe from 6.6–8 kya (Karelia HG, Samara HG, and to lesser degree Neolithic Motala HG and Hungary Gamba HG) (Supplemental Fig. S21F–Q) that predated the Yamnaya people. Mansi did not share the Late Upper Palaeolithic Caucasus HG ancestry with the Yamnaya people (Supplemental Fig. S25), suggesting that it is unlikely that Mansi have descended from the Yamnaya people. Therefore, Siberian admixtures into Northeastern Europe likely began prior to 6.6 kya, coinciding with the expansion of Y-Chromosome haplogroup N1c1 among Siberians and Northeastern Europeans (7.1–4.9 kya). Since haplogroup N likely originated in Asia (Shi et al. 2013) and currently achieves its highest frequency among Siberian populations, its presence among Eastern Europeans likely reflects ancient gene flows from Siberia into Northeastern Europe.
Genome Res. 2017. 27: 1-14
Genetic affinity between modern Siberians and Ust'-Ishim
To uncover ancient demographic events and reveal genetic links between present-day individuals and ancient ancestral populations, we compared 60 genomes from modern-day humans (Table 1; Supplemental Table S2) to 46 ancient genomes (Supplemental Table S3), including a 45,000-yr-old Siberian Ust’-Ishim individual (Fu et al. 2014), a 24,000-yr-old Siberian Mal'ta boy (MA-1), and a 17,000-yr-old Siberian individual (AG-2) from the Krasnoyarsk region (Raghavan et al. 2014).
Given that the bone from the Ust’-Ishim individual was found in Western Siberia, we reasoned that Ust’-Ishim might be related to modern Siberian populations. Indeed, this hypothesis was well supported by the TreeMix model (Fig. 6). We observed that the common ancestor of Siberians and East Asians traced 38% (95% CI: 28%–48%) of their ancestry to Ust’-Ishim's lineage (Fig. 6; Supplemental Fig. S14). In agreement with this, the D-statistics analysis (Fig. 5C; Supplemental Figs. S13C, S15) showed that Ust’-Ishim had higher genetic affinity with East Asians and Siberians, particularly Eastern Siberians, than with modern Europeans. This was also supported by the Y haplotype analysis. Previously, the ancient Siberian Ust’-Ishim (Fu et al. 2014) was thought to belong to the K2 Y-Chromosome clade, which is ancestral to haplogroup R (a Y-Chromosome clade extremely common in Europe), haplogroup Q (common among Native Americans and certain Siberians populations), haplogroup N (common in Siberia and Northeastern Europe), and haplogroup O (common in East Asia). However, we observed that Ust’-Ishim had a single derived SNP on Y-Chromosome specific to the NO branch (hg19 coordinate Chr Y 7,690,182) (Supplemental Fig. S12). This branch is associated with Siberians and some Northeastern European groups, as well as East Asians. Therefore, unlike previous claims, Ust’-Ishim's haplogroup most likely belongs to a more recent NO clade rather than to the more diverged K2 clade. Based on these lines of evidence, we conclude that Ust’-Ishim contributed more ancestry to the East Asian and Siberian populations than to modern Europeans or Native Americans.
Western Siberians are descendants of ANEs and Eastern Siberians
Our tree model grouped ANEs MA-1 and AG-2 with Western Siberians (Mansi and Nenets), suggesting a genetic link between ANEs and modern Western Siberians (Fig. 7). Furthermore, TreeMix estimated that 41% (95% CI: 36%–45%) of the ancestry of Andean Highlanders, the Native Americans, is attributable to the ANE lineage (Fig. 7; Supplemental Fig. S16). This is consistent with previous reports that Paleo-Americans trace ∼42% of their autosomal genome to an ancient Eurasian lineage related to MA-1 (Raghavan et al. 2014). We further tested these predictions using D-statistics by evaluating whether a modern individual shared significantly more derived alleles with MA-1 or AG-2 than with the East Asian Han individual (Fig. 5D; Supplemental Fig. S17A). Mansi, Khanty, Nenets, and Native Americans all had very strong ANE ancestry, demonstrated by their significant genetic affinities with MA-1 and AG-2. Therefore, Western Siberians share common ANE ancestry with Native Americans. The remaining ancestry of Western Siberians could be attributed to a population related to Eastern Siberians. The TreeMix model (Fig. 7) inferred that 43% of Mansi ancestry was derived from the admixture with a population related to Eastern Siberians Evenks and Evens, while 57% is attributable to the ANE ancestry. Therefore, genomes of Mansi people harbor significant ancestries from Siberian ANE people as well as Eastern Siberian populations in approximately equal proportions.

Figure 7.
Genetic relationships with ancient North Eurasians MA-1 and AG-2. Autosomal TreeMix admixture graph that includes ancient 7000-yr old La Brana (Spain) and low-coverage genomes of ancient Siberians MA-1 and AG-2. Residual plot for this graph is shown in Supplemental Figure S16. TreeMix model indicated multiple admixture events: 41% admixture (95% CI: 36%–45%) from the ANE clade represented by MA-1 and AG-2 individuals into Native Americans represented by Andean Highlanders, 43% admixture (95% CI: 38%–47%) between population related to Evenk and a common ancestor of Western Siberians (Mansi and Nenets), and 26% (95% CI: 16%–37%) admixture between population related to Even and modern-day Nenets. Additionally, Western Siberians and ANE are grouped together, indicating their shared genetic history.
To examine Siberian-related ancestry among ancient Europeans, we analyzed SNP data from the Holocene Eastern European hunter-gatherers (Haak et al. 2015) from the Samara and Karelia regions in modern Russia and the Motala region in Sweden (6.6–8 kya), as well as more recent Bronze Age Yamnaya samples (4.7–5.3 kya). D-Statistics demonstrated strong admixtures between the Mansi and nearly all hunter-gatherers from Eastern Europe (Supplemental Fig. S21), particularly for the Samara and Karelia samples. Slightly weaker, but significant affinities were present between Eastern Siberian Even and Eastern European hunter-gatherers. As previously mentioned, the time of the split of Eastern and Western Siberian populations was estimated to be around 6.8–9.9 kya. The weaker affinity of Eastern Siberians and Eastern European hunter-gatherers could be attributed to the Eastern Siberian ancestry of Mansi, which permeated into ancient European populations through Mansi-related admixture. Yamnaya samples also showed statistically significant admixtures with Mansi and Evens, but slightly weaker compared with Samara HG, which indicates a dilution of Eastern hunter-gatherer ancestry component of Yamnaya culture samples. The ANE-related ancestry among Eastern European hunter-gatherers could thus be attributed to gene flows between population ancestral to Mansi and Eastern European hunter-gatherers that occurred before 8 kya, as demonstrated by the strong genetic affinity between Mansi and Samara and Karelia hunter-gatherers from 6–8 kya.
Apart from Eastern European hunter-gatherers, Siberians also shared part of their ancestry with Pitted Ware Culture (PWC) 5,000-yr-old hunter-gatherers from Sweden Ire8 and Ajv52 (Skoglund et al. 2012). Ajv52 and particularly Ire8 had strong admixture signals with Western Siberians Mansi (Fig. 5E; Supplemental Fig. S17B), suggesting that like the closely related Yamnaya culture, they had strong ANE ancestries likely due to admixtures with Mansi-related population.
Shared ancestry between Eastern Siberians and ancient human of the Saqqaq culture
Our data also showed evidence of Eastern Siberian ancestry within the genome of a 4000-yr-old Saqqaq individual (Rasmussen et al. 2010) from Greenland. The tree model with Saqqaq genome showed that Saqqaq was related to both East Asians and Eastern Siberians, and 12% of its ancestry was attributable to a population related to modern-day Evens (Supplemental Figs. S19, S20). The detected gene flow was strongly supported by the D-statistics (Supplemental Figs. S7F, S8F). In addition, 18% of American Andeans' ancestry came from Evens (Supplemental Fig. S19). This, together with the substantial gene flow from an ancestor of Evens and Evenks to Altayans, suggests that the signal from Evens to Saqqaq can be a result of ancient admixture happened between the ancestor of Siberians (Eastern Siberians) and early migrants to the Americas.

Figure 8.
Geographical dispersion model. The approximate model of gene flows reflecting divergence of primary clades in Europe, East Asia, and Siberia with approximate divergence times based on Chromosome Y, MSMC, TreeMix, and D-statistics analyses. Percentages represent proportions of ancestry contributed to each lineage from other lineages. The common ancestors of Mansi, Khanty, and Nenets had derived 57% of their ancestry from an ANE population, related to MA-1, AG-2, and Native Americans and 43% from the admixture with a population related to Evens and Evenks. Native Americans trace 42% of their ancestry to ANE and 58% to a common ancestor of Eastern Siberians and East Asians. In the figure, 45 kya represents the split between Y-Chromosome clades QR and NO; 33 kya represents the split between Y-Chromosome Q and R clades and mtDNA N2 and W clades; 17 kya represents Mansi-Andean separation time; 44 kya represents the split of Ust’-Ishim's haplogroup from NO clade; 19 kya represents Han-Andean separation time; 10 kya represents Han-Eastern Siberian separation time; and 5.3–9.9 kya represents Evenk-Mansi MSMC separation time (6.8–9.9 kya) and expansion of N1c1 Y-haplogroup (5.3–7.1 kya). In this figure, Europeans represent a collective term for the following populations from this study: Komi, Veps, Karelians, Russians, and Belarussians.
Discussion
Genomes of indigenous Siberians harbor evidence of many important ancient events that shaped population history and peopling of Eurasia and the New World. Siberia is home to more than 30 ethnic groups that were thus far undersurveyed in population genetic studies. In this work, we examined the genetic history of several Siberian and Eastern European populations and analyzed their genetic links to ancient and modern populations. We summarized our findings in the form of a synthetic geographical dispersion model (Fig. 8), which describes how populations and genetic variation likely spread across Northern Eurasia.
Our analyses demonstrated that both East Asian and Siberian populations shared a significant amount of ancestry (38%) attributable to the ancient Siberian Ust’-Ishim individual, thus pointing to very ancient origins of indigenous Siberians. D-Statistics tests also suggested stronger affinity of Ust’-Ishim to both Eastern and Western Siberians (Fig. 5C; Supplemental Figs. S13C, S15) than to modern Europeans. Another line of evidence pointing to affinity of both East Asians and Eastern Siberians with Ust’-Ishim was provided by the Y-Chromosome haplogroup analysis (Fig. 4A; Supplemental Fig. S12). We showed that Ust’-Ishim's Y-Chromosome haplogroup belonged to NO clade, and its divergence from NO lineage predated the split of Y-Chromosome haplogroup N (particularly common in Siberia and Northeastern Europe) and haplogroup O (the most common Y-Chromosome haplogroup among Eastern and Southern Asian populations). These new relationships were uncovered largely due to the inclusion of the Western and Eastern Siberian genomes, including Mansi, Khanty, Nenets, Evens, Buryat, and other Siberian individuals. Therefore, the new findings described here provide a more accurate interpretation of patterns observed in the original Ust’-Ishim study (Fu et al. 2014).
A particularly surprising finding was the genetic link between 24,000-yr-old Siberian Mal'ta boy, Native Americans, and the Western Siberians Mansi, Khanty, and Nenets. This finding demonstrates a strong genetic link between Western Siberians and Native Americans due to their common ANE ancestry. We estimate that 57% of Western Siberian Mansi and Khanty ancestry was related to the ANE lineage represented by MA-1 and AG-2 ancient individuals. The other important finding was that the remaining 43% of Western Siberian ancestry was related to Eastern Siberian populations, suggesting admixtures between Eastern Siberian groups most related to Evens and Evenks and the common ancestors of Western Siberians (Fig. 2). This is particularly interesting in the context of ancestral relationships of modern Northeastern European populations. Lazaridis et al. (2014) modeled the ancestry of Eastern Europeans such as Mordovians, Finns, Russians, Saami, and Chuvashs but could not completely explain their genetic makeup by a mixture of three early European groups: the ancient northern Eurasians (represented by MA-1), the West European hunter-gatherers (represented by Loschbour), and the early European farmers (represented by Stuttgart). This was proposed to be due to a greater relatedness of Eastern Europeans to East Asians, compared with other European groups. However, our analyses of genetic relationships of Northeastern European populations such as Mezen Russians, Komi, Karelians, and Veps suggested that the unexplained ancestral components among modern Europeans likely resulted from their affinity with both Western and Eastern Siberian populations.
Our findings also showed that Western Siberians Mansi and Khanty and ancient Eastern European populations (Yamnaya and the PWC people) shared a significant amount of ANE-related ancestry. The genetic affinity of ancient Eastern Europeans with Eastern Siberians is relatively weaker (Fig. 5E; Supplemental Figs. S21, S24). Therefore, we propose that the Western Siberian admixture into Northeastern Europeans likely began before the Yamnaya culture period (5.3–4.7 kya), since the admixtures with Mansi are also very strong among hunter-gatherers from Northeastern Europe from 6.6–8 kya (Karelia HG, Samara HG, and to lesser degree Neolithic Motala HG and Hungary Gamba HG) (Supplemental Fig. S21F–Q) that predated the Yamnaya people. Mansi did not share the Late Upper Palaeolithic Caucasus HG ancestry with the Yamnaya people (Supplemental Fig. S25), suggesting that it is unlikely that Mansi have descended from the Yamnaya people. Therefore, Siberian admixtures into Northeastern Europe likely began prior to 6.6 kya, coinciding with the expansion of Y-Chromosome haplogroup N1c1 among Siberians and Northeastern Europeans (7.1–4.9 kya). Since haplogroup N likely originated in Asia (Shi et al. 2013) and currently achieves its highest frequency among Siberian populations, its presence among Eastern Europeans likely reflects ancient gene flows from Siberia into Northeastern Europe.
Genome Res. 2017. 27: 1-14




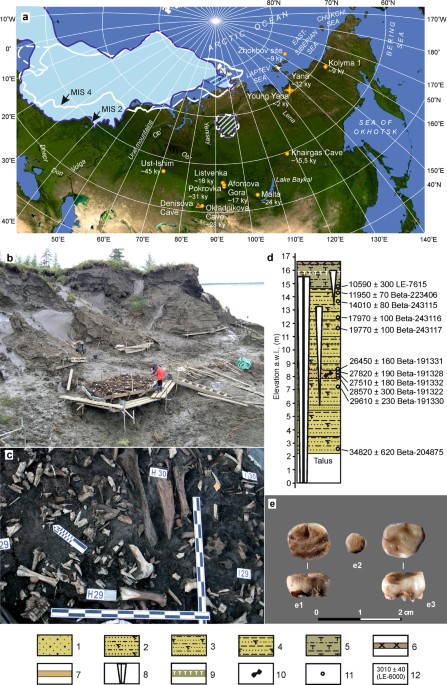

 raphic locations for the USR1 and NNA–SNA splits.
raphic locations for the USR1 and NNA–SNA splits.