|
|
Post by Admin on Apr 29, 2018 18:40:17 GMT
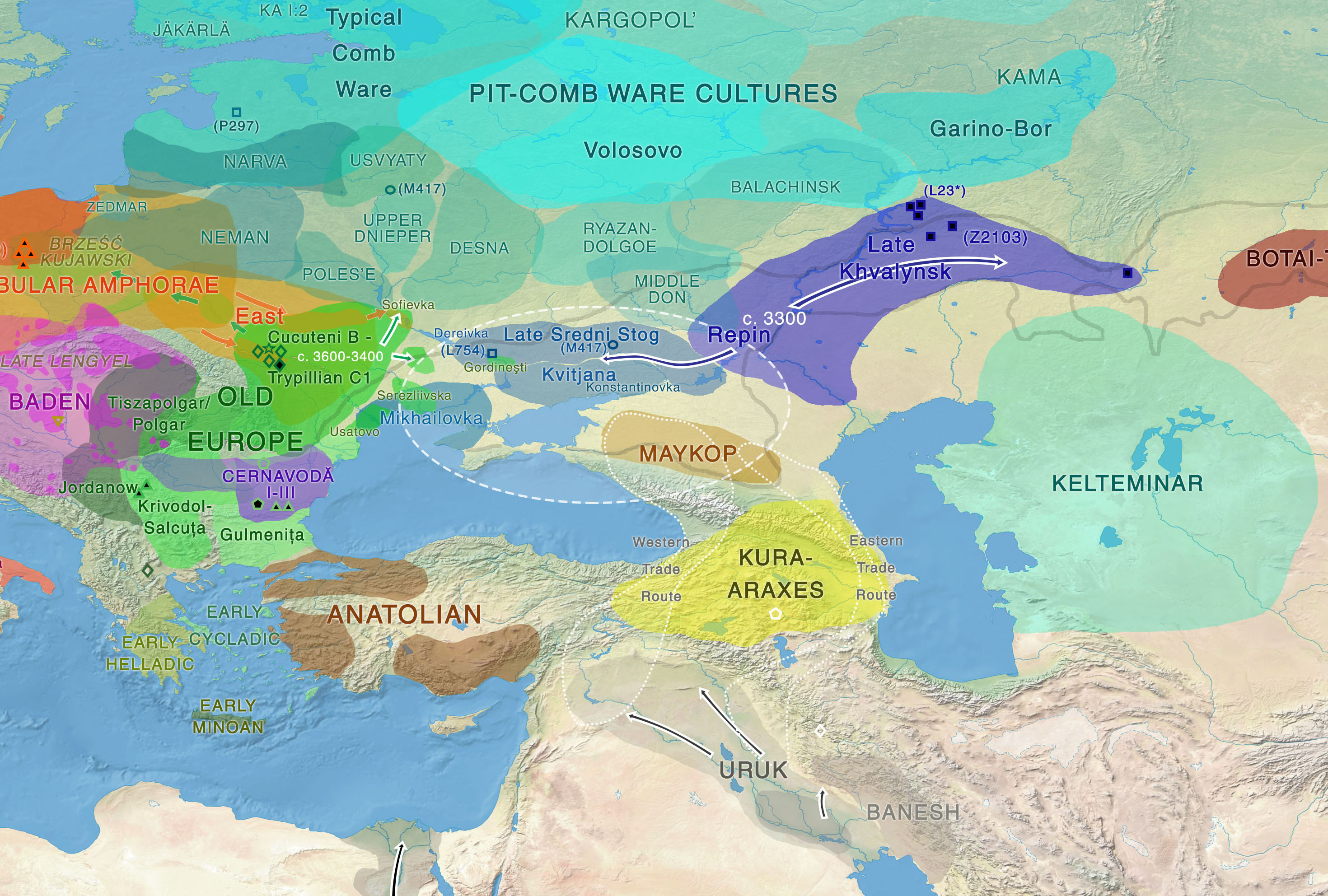
Yuri Y. Rassamakin - The Maikop-Novosvobodnaia Culture Development The Latest Eneolithic -- Early Bronze Age of the Black Sea Steppe in the Context of the Maikop-Novosvobodnaia Culture Development (the Second Half of the 4th Millennium BC)(session 2)  It's been theorized that R1b people from the Middle East migrated across the Caucasus and established the Maikop culture, before expanding throughout the Pontic-Caspian steppes and mixing with the indigenous R1a steppe people. The Proto-Indo-European language was brought by the more advanced and dominant partner (R1b) and adopted by the R1a population at the same time along with the rest of the cultural elements from Maikop. The Maikop culture was the first kurgan culture established between the Russian steppes and Mesopotamia, which was also adjacent to the Yamnaya culture. As David Reich explains in his book "Who We Are and How We Got Here" (2018, p.109), the penetration of Maikop lands by some people of Iranian-related ancestry is plausible because Maikop goods were heavily influenced by the Uruk civilization. 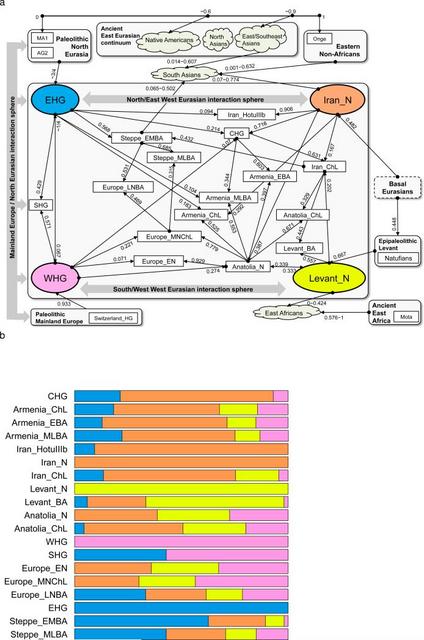 Modelling ancient West Eurasians, East Africans, East Eurasians and South Asians Lazaridis et al. (2017) showed that the Yamnaya, tagged as Steppe EMBA, are best modeled as a mixture of Eastern European Hunter-Gatherers (EHG) and Chalcolithic farmers from western Iran. The mixture ratios are 56.8/43.2, respectively. The migration proceeded via the Caucasus isthmus between the Black and Caspian Seas and this type of migration continued up until the time of the Maikop culture, which preceded the Yamnaya culture. Haak et al. (2015) also found that Y chromosome Yamnaya haplogroups were R1b1a or R1b1a2. Another recent study by Mathieson et al. (2015) supports this result, though claims affinity with an “Armenian-like Near Eastern source.” 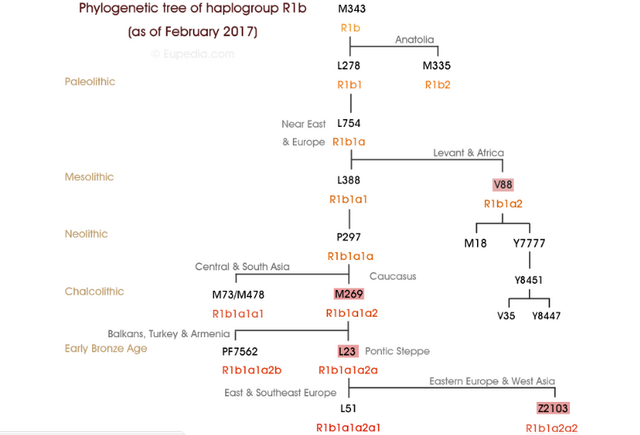 Haplogroup R1b-M269 (R1b1a1a2) was found at a substantially high frequency (15%) in the northern portion of the Iranian Plateau (Terreros et al. 2011). Haplogroup R1b-M269 shows its highest frequency in the Assyrians (29.2%, averaged on Tehran and Azerbaijan Gharbi groups) and high frequencies were observed (∼24%) in the Armenians from Tehran and in Lorestan (Grugni et al. 2012). Its descendant subclade R1b-Z2103 (R1b1a2a2) has been found to be prevalent in ancient DNA associated with the Yamnaya culture. 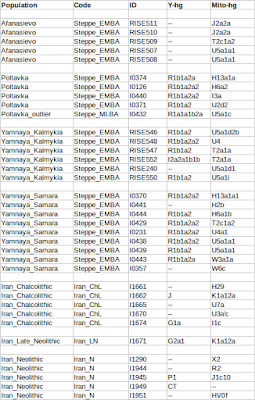 According to the Indo-Hittite hypothesis, the Anatolian languages may have split off a Pre-Proto-Indo-European language considerably earlier than the separation of the remaining Indo-European languages. It's likely that the ancient Hittites already spoke a Pre-Proto-Indo-European language prior to the time of Yamnaya culture and Yamnaya steppe herders were not responsible for the diffusion of IE languages to Anatolia. Tocharian has the perfect wagon vocabulary but some key words are missing in Indo-Hittite, which preserves archaism lost in other IE languages. Probably the ancient Hittites didn't develop the wagon vocabulary because they stayed behind unlike Tocharians who migrated to the Tarim Basin in western China. 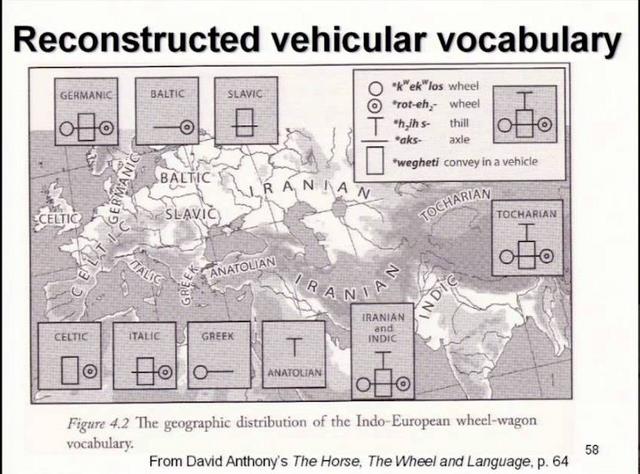 "First, the lack of genetic indications for an intrusion into Anatolia refutes the classical notion of a Yamnaya-derived mass invasion or conquest. However, it does fit the recently developed consensus among linguists and historians that the speakers of the Anatolian languages established themselves in Anatolia by gradual infiltration and cultural assimilation. Our findings corroborate the Indo-Anatolian Hypothesis, which claims that Anatolian Indo-European split off from Proto-Indo-European first and that Anatolian Indo-European represents a sister rather than a daughter language. Our findings call for theidentification of the speakers of Proto-Indo-Anatolian as a population earlier that the Yamnaya and late Maykop cultures." (Linguistic supplement to Damgaard et al. 2018: Early Indo-European languages, Anatolian, Tocharian and Indo-Iranian) Abstract The North Caucasus region is rich in early Bronze Age sites, with burials yielding many artifacts, including those from the Chekon, Natukhaevskaya, Katusvina-Krivitsa kurgan groups (at Krasnodar Krai, Russia) and Klady kurgan (near Novosvobodnaya Village, Republic of Adygea, Russia). According to the mainstream archaeological hypothesis, these sites belong to the Maikop culture (3700–3000 years BC), with Novosvobodnaya communities representing an offshoot of Maikop ancestry. However, due to specific differences in Novosvobodnaya artifacts, the Maikop and Novosvobodnaya assemblages could represent two synchronous archaeological cultures living in almost sympatry but showing independent ancestry, from the Near East and Europe respectively.  Here, we used target-enrichment together with high-throughput sequencing to characterize the complete mitochondrial sequence of three Maikop and three Novosvobodnaya individuals. We identified T2b, N1b1 and V7 haplogroups, all widely spread in Neolithic Europe. In addition, we identified the Paleolithic Eurasian U8b1a2 and M52 haplogroups, which are frequent in modern South Asia, particularly in modern India. Our data provide a deeper understanding of the diversity of Early Bronze Age North Caucasus communities and hypotheses of its origin. Analyzing non-human sequencing reads for microbial content, we found that one individual from the Klady kurgan was infected by the pathogen Brucella abortus that is responsible for zoonotic infections from cattle to humans. This finding is in agreement with Maikop/Novosvobodnaya livestock groups, mostly consisting of domestic pigs and cattle. This paper represents a first mitochondrial genome analysis of Maikop/Novosvobodnaya culture as well as the earliest brucellosis case in archaeological humans. Journal of Archaeological Science Volume 73, September 2016, Pages 138-144 |
|
|
|
Post by Admin on Apr 30, 2018 18:45:35 GMT
 Between 10,000-9,000 BCE, humans began practicing agriculture in the Near East1. In the ensuing five millennia, plants and animals domesticated in the Near East spread throughout West Eurasia (a vast region that also includes Europe) and beyond. The relative homogeneity of present-day West Eurasians in a world context2 suggests the possibility of extensive migration and admixture that homogenized geographically and genetically disparate sources of ancestry. The spread of the world's first farmers from the Near East would have been a mechanism for such homogenization. To date, however, due to the poor preservation of DNA in warm climates, it has been impossible to study the population structure and history of the first farmers and to trace their contribution to later populations. In order to overcome the obstacle of poor DNA preservation, we took advantage of two methodological developments. First, we sampled from the inner ear region of the petrous bone3,4 that can yield up to ~100 times more endogenous DNA than other skeletal elements4. Second, we used in-solution hybridization5 to enrich extracted DNA for about 1.2 million single nucleotide polymorphism (SNP) targets6,7, making efficient sequencing practical by filtering out microbial and non-informative human DNA. We merged all sequences extracted from each individual, and randomly sampled a single sequence with minimum mapping and sequence quality to represent each SNP, restricting to individuals with at least 9,000 SNPs covered at least once (Methods). We obtained genome-wide data passing quality control for 45 individuals on whom we had a median coverage of 172,819 SNPs. We assembled radiocarbon dates for 26 individuals (22 new generated for this study) (Supplementary Data Table 1). The newly reported ancient individuals date to ~12,000-1,400 BCE and come from the southern Caucasus (Armenia), northwestern Anatolia (Turkey), Iran, and the southern Levant (Israel and Jordan) (Supplementary Data Table 1, Fig. 1a). (One individual had a radiocarbon date that was not in agreement with the date of its archaeological context and was also a genetic outlier.) The samples include Epipaleolithic Natufian hunter-gatherers from Raqefet Cave in the Levant (12,000-9,800 BCE); a likely Mesolithic individual from Hotu Cave in the Alborz mountains of Iran (probable date of 9,100-8,600 BCE); Pre-Pottery Neolithic farmers from ‘Ain Ghazal and Motza in the southern Levant (8,300-6,700 BCE); and early farmers from Ganj Dareh in the Zagros mountains of western Iran (8,200-7,600 BCE). The samples also include later Neolithic, Chalcolithic (~4,800-3,700 BCE), and Bronze Age (~3,350-1,400 BCE) individuals (Supplementary Information, section 1). We combined our data with previously published ancient data7,8,9,10,8,10-15 to form a dataset of 281 ancient individuals. We then further merged with 2,583 present-day people genotyped on the Affymetrix Human Origins array13,16 (238 new) (Supplementary Data Table 2; Supplementary Information, section 2). We grouped the ancient individuals based on archaeological culture and chronology (Fig. 1a; Supplementary Data Table 1). We refined the grouping based on patterns evident in Principal Components Analysis (PCA)17 (Fig. 1b; Extended Data Fig. 1), ADMIXTURE model-based clustering18 (Extended Data Fig. 2a), and ‘outgroup’ f3-analysis (Extended Data Fig. 3). We used f4-statistics to identify outlier individuals and to cluster phylogenetically indistinguishable groups into ‘Analysis Labels’ (Supplementary Information, section 3). 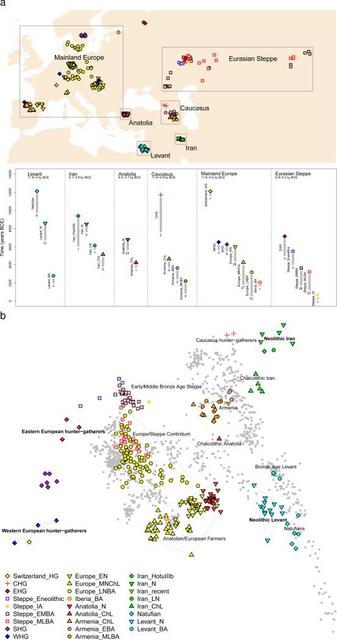 Figure 1 Genetic structure of ancient West Eurasia (a) Sampling locations and times in six regions. Sample sizes for each population are given below each bar. Abbreviations used: E: Early, M: Middle, L: Late, HG: Hunter-Gatherer, N: Neolithic, ChL: Chalcolithic, BA: Bronze Age, IA: Iron Age. (b) Principal components analysis of 991 present-day West Eurasians (grey points) with 278 projected ancient samples (excluding the Upper Paleolithic Ust’-Ishim, Kostenki14, and MA1). To avoid visual clutter, population labels of present-day individuals are shown in Extended Data Fig. 1. We analyzed these data to address six questions. (1) Previous work has shown that the first European farmers harboured ancestry from a Basal Eurasian lineage that diverged from the ancestors of north Eurasian hunter-gatherers and East Asians before they separated from each other13. What was the distribution of Basal Eurasian ancestry in the ancient Near East? (2) Were the first farmers of the Near East part of a single homogeneous population, or were they regionally differentiated? (3) Was there continuity between late pre-agricultural hunter-gatherers and early farming populations, or were the hunter-gatherers largely displaced by a single expansive population as in early Neolithic Europe?8 (4) What is the genetic contribution of these early Near Eastern farmers to later populations of the Near East? (5) What is the genetic contribution of the early Near Eastern farmers to later populations of mainland Europe, the Eurasian steppe, and to populations outside West Eurasia? (6) Do our data provide broader insights about population transformations in West Eurasia? 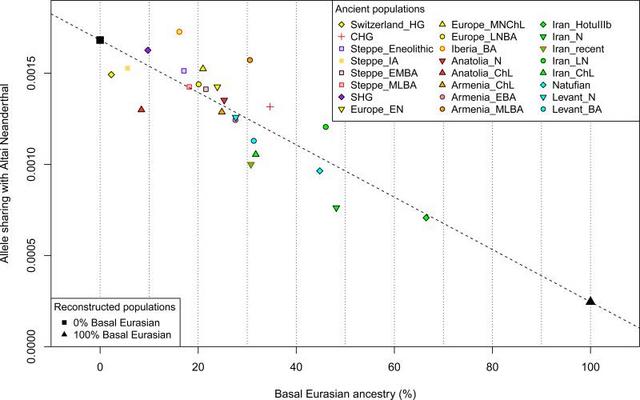 Figure 2 Basal Eurasian ancestry explains the reduced Neanderthal admixture in West Eurasians Basal Eurasian ancestry estimates are negatively correlated to a statistic measuring Neanderthal ancestry f4(Test, Mbuti; Altai, Denisovan). West Eurasians harbour significantly less Neanderthal ancestry than East Asians19-21, which could be explained if West Eurasians (but not East Asians) have partial ancestry from a source diluting their Neanderthal inheritance22. Supporting this theory, we observe a negative correlation between Basal Eurasian ancestry and the rate of shared alleles with Neanderthals19 (Supplementary Information, section 5; Fig. 2). By extrapolation, we infer that the Basal Eurasian population had lower Neanderthal ancestry than non-Basal Eurasian populations and possibly none (ninety-five percent confidence interval truncated at zero of 0-60%; Fig. 2; Methods). The finding of little if any Neanderthal ancestry in Basal Eurasians could be explained if the Neanderthal admixture into modern humans 50,000-60,000 years ago11 largely occurred after the splitting of the Basal Eurasians from other non-Africans. It is striking that the highest estimates of Basal Eurasian ancestry are from the Near East, given the hypothesis that it was there that most admixture between Neanderthals and modern humans occurred19,23. This could be explained if Basal Eurasians thoroughly admixed into the Near East before the time of the samples we analyzed but after the Neanderthal admixture. Alternatively, the ancestors of Basal Eurasians may have always lived in the Near East, but the lineage of which they were a part did not participate in the Neanderthal admixture. A population without Neanderthal admixture, basal to other Eurasians, may have plausibly lived in Africa. Craniometric analyses have suggested an affinity between the Natufians and populations of north or sub-Saharan Africa24,25, a result that finds some support from Y chromosome analysis which shows that the Natufians and successor Levantine Neolithic populations carried haplogroup E, of likely ultimate African origin, which has not been detected in other ancient males from West Eurasia (Supplementary Information, section 6) 7,8. However, no affinity of Natufians to sub-Saharan Africans is evident in our genome-wide analysis, as present-day sub-Saharan Africans do not share more alleles with Natufians than with other ancient Eurasians (Extended Data Table 1). (We could not test for a link to present-day North Africans, who owe most of their ancestry to back-migration from Eurasia26,27.) The idea of Natufians as a vector for the movement of Basal Eurasian ancestry into the Near East is also not supported by our data, as the Basal Eurasian ancestry in the Natufians (44±8%) is consistent with stemming from the same population as that in the Neolithic and Mesolithic populations of Iran, and is not greater than in those populations (Supplementary Information, section 4). Further insight into the origins and legacy of the Natufians could come from comparison to Natufians from additional sites, and to ancient DNA from north Africa. |
|
|
|
Post by Admin on May 1, 2018 18:24:04 GMT
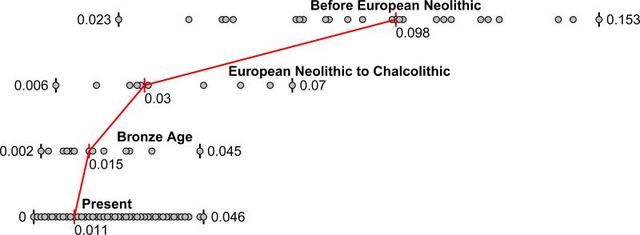 Figure 3 Genetic differentiation and its dramatic decrease over time in West Eurasia Pairwise FST distribution among populations belonging to four successive time slices in West Eurasia; the median (red) and range of FST is shown. PCA on present-day West Eurasian populations (Methods) (Extended Data Fig. 1) on which we projected the ancient individuals (Fig. 1b) replicates previous findings of a Europe-Near East contrast along the horizontal Principal Component 1 (PC1) and parallel clines (PC2) in both Europe and the Near East (Extended Data Fig. 1)7,8,13. Ancient samples from the Levant project at one end of the Near Eastern cline, and ancient samples from Iran at the other. The two Caucasus Hunter Gatherers (CHG)9 are less extreme along PC1 than the Mesolithic and Neolithic individuals from Iran, while individuals from Chalcolithic Anatolia, Iran, and Armenia, and Bronze Age Armenia occupy intermediate positions. Qualitatively, the PCA has the appearance of a quadrangle whose four corners are some of the oldest samples: bottom-left: Western Hunter Gatherers (WHG), top-left: Eastern Hunter Gatherers (EHG), bottom-right: Neolithic Levant and Natufians, top-right: Neolithic Iran. This suggests the hypothesis that diverse ancient West Eurasians can be modelled as mixtures of as few as four streams of ancestry related to these populations, which we confirmed using qpWave7 (Supplementary Information, section 7). We computed squared allele frequency differentiation between all pairs of ancient West Eurasians28 (Methods; Fig. 3; Extended Data Fig. 2b; Extended Data Fig. 4), and found that the populations at the four corners of the quadrangle had differentiation of FST=0.08-0.15, comparable to the value of 0.09-0.13 seen between present-day West Eurasians and East Asians (Han) (Supplementary Data Table 3). In contrast, by the Bronze Age, genetic differentiation between pairs of West Eurasian populations had reached its present-day low levels (Fig. 3): today, FST is ≤0.025 for 95% of the pairs of West Eurasian populations and ≤0.046 for all pairs (Fig. 3). These results point to a demographic process that established high differentiation across West Eurasia and then reduced this differentiation over time.  Figure 4 Modelling ancient West Eurasians, East Africans, East Eurasians and South Asians (a) All the ancient populations can be modelled as mixtures of two or three other populations and up to four proximate sources (marked in colour). Mixture proportions inferred by qpAdm are indicated by the incoming arrows to each population. Clouds represent sets of more than one population. Multiple admixture solutions are consistent with the data for some populations, and while only one solution is shown here, Supplementary Information, section 7 presents the others. (b) A flat representation of the graph showing mixture proportions from the four proximate sources. Almost all ancient and present-day West Eurasians have evidence of significant admixture between two or more ancestral populations, as documented by statistics of the form f3(Test; Reference1, Reference2) which if negative, show that a Test population's allele frequencies tend to be intermediate between two Reference populations16 (Extended Data Table 2). To better understand the admixture history beyond these patterns, we used qpAdm7, which can evaluate whether a particular Test population is consistent with being derived from a set of proposed source populations, and if so, infer mixture proportions (Methods). We used this approach to carry out a systematic survey of ancient West Eurasian populations to explore their possible sources of admixture (Fig. 4; Supplementary Information, section 7). Among first farmers, those of the Levant trace ~2/3 of their ancestry to people related to Natufian hunter-gatherers and ~1/3 to people related to Anatolian farmers (Supplementary Information, section 7). Western Iranian first farmers cluster with the likely Mesolithic HotuIIIb individual and more remotely with hunter-gatherers from the southern Caucasus (Fig. 1b), and share alleles at an equal rate with Anatolian and Levantine early farmers (Supplementary Information, section 7), highlighting the long-term isolation of western Iran.  During subsequent millennia, the early farmer populations of the Near East expanded in all directions and mixed, as we can only model populations of the Chalcolithic and subsequent Bronze Age as having ancestry from two or more sources. The Chalcolithic people of western Iran can be modelled as a mixture of the Neolithic people of western Iran, the Levant, and Caucasus Hunter Gatherers (CHG), consistent with their position in the PCA (Fig. 1b). Admixture from populations related to the Chalcolithic people of western Iran had a wide impact, consistent with contributing ~44% of the ancestry of Levantine Bronze Age populations in the south and ~33% of the ancestry of the Chalcolithic northwest Anatolians in the west. Our analysis show that the ancient populations of the Chalcolithic Iran, Chalcolithic Armenia, Bronze Age Armenia and Chalcolithic Anatolia were all composed of the same ancestral components, albeit in slightly different proportions (Fig. 4b; Supplementary Information, section 7). Admixture did not only occur within the Near East but extended towards Europe. To the north, a population related to people of the Iran Chalcolithic contributed ~43% of the ancestry of early Bronze Age populations of the steppe. The spread of Near Eastern ancestry into the Eurasian steppe was previously inferred7 without access to ancient samples, by hypothesizing a population related to present-day Armenians as a source7,8. To the west, the early farmers of mainland Europe were descended from a population related to Neolithic northwestern Anatolians8. This is consistent with an Anatolian origin of farming in Europe, but does not reject other sources, since the spatial distribution of the Anatolian/European-like farmer populations is unknown. We can rule out the hypothesis that European farmers stem directly from a population related to the ancient farmers of the southern Levant29,30, however, since they share more allele with Anatolian Neolithic farmers than with Levantine farmers as attested by the positive statistic f4(Europe_EN, Chimp; Anatolia_N, Levant_N) (Z=15). Migrations from the Near East also occurred towards the southwest into East African populations which experienced West Eurasian admixture ~1,000 BCE31,32. Previously, the West Eurasian population known to be the best proxy for this ancestry was present-day Sardinians32, who resemble Neolithic Europeans genetically13,33. However, our analysis shows that East African ancestry is significantly better modelled by Levantine early farmers than by Anatolian or early European farmers, implying that the spread of this ancestry to East Africa was not from the same group that spread Near Eastern ancestry into Europe (Extended Data Fig. 5; Supplementary Information, section 8). Nature. 2016 Aug 25; 536(7617): 419–424. |
|
|
|
Post by Admin on May 2, 2018 18:56:08 GMT
 Figure 1: Genetic structure of ancient Europe. Deep coalescence of early Holocene lineages The geographical proximity of the Southern Caucasus to the Levant begs the question of whether CHG might be related to early Neolithic farmers with Near Eastern heritage. To address this question formally we reconstructed the relationship among WHG, CHG and EF using available high-quality ancient genomes1,3. We used outgroup f3-statistics14 to compare the three possible topologies, with the correct relationship being characterized by the largest amount of shared drift between the two groups that form a clade with respect to the outgroup (Fig. 2a; Supplementary Note 4). A scenario in which the population ancestral to both CHG and EF split from WHG receives the highest support, implying that CHG and EF form a clade with respect to WHG. We can reject a scenario in which CHG and WHG form a distinct clade with respect to EF. The known admixture of WHG with EF1,3,4,5 implies that some shared drift is found between WHG and EF with respect to CHG, but this is much smaller than the shared drift between CHG and EF. Thus, WHG split first, with CHG and EF separating only at a later stage.  Figure 2: The relationship between Caucasus hunter-gatherers (CHG), western hunter-gatherers and early farmers. We next dated the splits among WHG, CHG and EF using a coalescent model implemented with G-PhoCS15 based on the high-coverage genomes in our data set (Fig. 2b for a model using the German farmer Stuttgart1 to represent EF; and Supplementary Table 5 for models using the Hungarian farmer NE1 (ref. 3)) and taking advantage of the mutation rate recently derived from Ust’-Ishim10. G-Phocs dates the split between WHG and the population ancestral to CHG and EF at ∼40–50 kya (range of best estimates depending on which genomes are used; see Supplementary Table 5 and Supplementary Note 5 for details), implying that they diverged early on during the colonisation of Europe16, and well before the LGM. On the WHG branch, the split between Bichon and Loschbour1 is dated to ∼16–18 kya (just older than the age of Bichon), implying continuity in western Europe, which supports the conclusions from our previous analyses. The split between CHG and EF is dated at ∼20–30 kya emerging from a common basal Eurasian lineage1 (Supplementary Fig. 2) and suggesting a possible link with the LGM, although the broad confidence intervals require some caution with this interpretation. In any case, the sharp genomic distinctions between these post-LGM populations contrasts with the comparative lack of differentiation between the earlier Eurasian genomes, for example, as visualized in the ADMIXTURE analysis (Fig. 1a), and it seems likely that this structure emerged as a result of ice age habitat restriction. Like EF, but in contrast to WHG, CHG carry a variant of the SLC24A5 gene17 associated with light skin colour (rs1426654, see Supplementary Note 6). This trait, which is believed to have risen to high frequency during the Neolithic expansion18, may thus have a relatively long history in Eurasia, with its origin probably predating the LGM. A partial genome from a 24,000-year-old individual (MA1) from Mal’ta, Siberia6 had been shown to be divergent from other ancient samples and was shown by Lazaridis et al.1, using f4 statistics, to have more shared alleles with nearly all modern Europeans than with an EF genome. This allowed inference of an ANE component in European ancestry, which was subsequently shown to have an influence in later eastern hunter-gatherers and to have spread into Europe via an incursion of Steppe herders beginning ∼4,500 years ago5,7. Several analyses indicate that CHG genomes are not a subset of this ANE lineage. First, MA1 and CHG plot in distinct regions of the PCA and also have very different profiles in the ADMIXTURE analysis (Fig. 1). Second, when we test if CHG shows any evidence of excess allele sharing with MA1 relative to WHG using tests of the form D(Yoruba, CHG; MA1, WHG) no combinations were significantly positive (Supplementary Table 6). Last, we also tested whether the ancestral component inferred in modern Europeans from MA1 was distinct from any that may have been donated from CHG using tests of the form D(Yoruba, MA1; CHG, modern North European population) (Supplementary Table 7). All northern Europeans showed a significant sharing of alleles with MA1 separate to any they shared with CHG. |
|
|
|
Post by Admin on May 3, 2018 18:31:04 GMT
 Figure 3: Distribution of ROH. Caucasus hunter-gatherer contribution to subsequent populations We next explored the extent to which Bichon and CHG contributed to contemporary populations using outgroup f3(African; modern, ancient) statistics, which measure the shared genetic history between an ancient genome and a modern population since they diverged from an African outgroup. Bichon, like younger WHG, shows strongest affinity to northern Europeans (Supplementary Fig. 3), while contemporary southern Caucasus populations are the closest to CHG (Fig. 4a and Supplementary Fig. 3), thus implying a degree of continuity in both regions stretching back at least 13,000 years to the late Upper Palaeolithic. Continuity in the Caucasus is also supported by the mitochondrial and Y chromosomal haplogroups of Kotias (H13c and J2a, respectively) and Satsurblia (K3 and J), which are all found at high frequencies in Georgia today22,23,24 (Supplementary Note 8).  Figure 4: The relationship of Caucasus hunter-gatherers to modern populations. CHG origins of migrating Early Bronze Age herders
We investigated the temporal stratigraphy of CHG influence by comparing these data to previously published ancient genomes. We find that CHG, or a population close to them, contributed to the genetic makeup of individuals from the Yamnaya culture, which have been implicated as vectors for the profound influx of Pontic steppe ancestry that spread westwards into Europe and east into central Asia with metallurgy, horseriding and probably Indo-European languages in the third millenium BC5,7. CHG ancestry in these groups is supported by ADMIXTURE analysis (Fig. 1b) and admixture f3-statistics14,25 (Fig. 5), which best describe the Yamnaya as a mix of CHG and Eastern European hunter-gatherers. The Yamnaya were semi-nomadic pastoralists, mainly dependent on stock-keeping but with some evidence for agriculture, including incorporation of a plow into one burial26. As such it is interesting that they lack an ancestral coefficient of the EF genome (Fig. 1b), which permeates through western European Neolithic and subsequent agricultural populations. During the Early Bronze Age, the Caucasus was in communication with the steppe, particularly via the Maikop culture27, which emerged in the first-half of the fourth millennium BC. The Maikop culture predated and, possibly with earlier southern influences, contributed to the formation of the adjacent Yamnaya culture that emerged further to the north and may be a candidate for the transmission of CHG ancestry. In the ADMIXTURE analysis of later ancient genomes (Fig. 1b) the Caucasus component gives a marker for the extension of Yamnaya admixture, with substantial contribution to both western and eastern Bronze Age samples. However, this is not completely coincident with metallurgy; Copper Age genomes from Northern Italy and Hungary show no contribution; neither does the earlier of two Hungarian Bronze Age individuals.The separation between CHG and both EF and WHG ended during the Early Bronze Age when a major ancestral component linked to CHG was carried west by migrating herders from the Eurasian Steppe. The foundation group for this seismic change was the Yamnaya, who we estimate to owe half of their ancestry to CHG-linked sources. These sources may be linked to the Maikop culture, which predated the Yamnaya and was located further south, closer to the Southern Caucasus. Through the Yamanya, the CHG ancestral strand contributed to most modern European populations, especially in the northern part of the continent. Nature Communications volume 6, Article number: 8912 (2015) |
|















