|
|
Post by Admin on Jul 10, 2018 18:55:30 GMT
 Figure 2: PC analysis based on mitochondrial DNA haplogroup frequencies. Mitochondrial lineages in the NPR Scythians analyzed in this study appear to consist of a mixture of west and east Eurasian haplogroups. West Eurasian lineages were represented by subdivisions of haplogroup U5 (U5a2a1, U5a1a1, U5a1a2b, U5a2b, U5a1b, U5b2a1a2, six individuals total, 31.6%), H (H and H5b, three individuals total, 15.8%), J (J1c2 and J2b1a6, two individuals, 10.5%), as well as haplogroups N1b1a, W3a and T2b (one individual each, 5.3% each specimen). East Eurasian mt lineages were represented by haplogroups A, D4j2, F1b, M10a1a1a, and H8c (represented by a single individual), in total, comprising 26.3% of our sample set. The results of the low-resolution mtDNA screening at GVSU were consistent with whole mt capture results from AMU (see Supplementary Table S1). Polymorphism patterns uncovered in the coding and HVRI regions in specimens SCY009 and SCY011 identified their lineages to belong to haplogroups J and A respectively. Specimens SCY006 and SCY010 were assigned to the M* and N* clades respectfully. 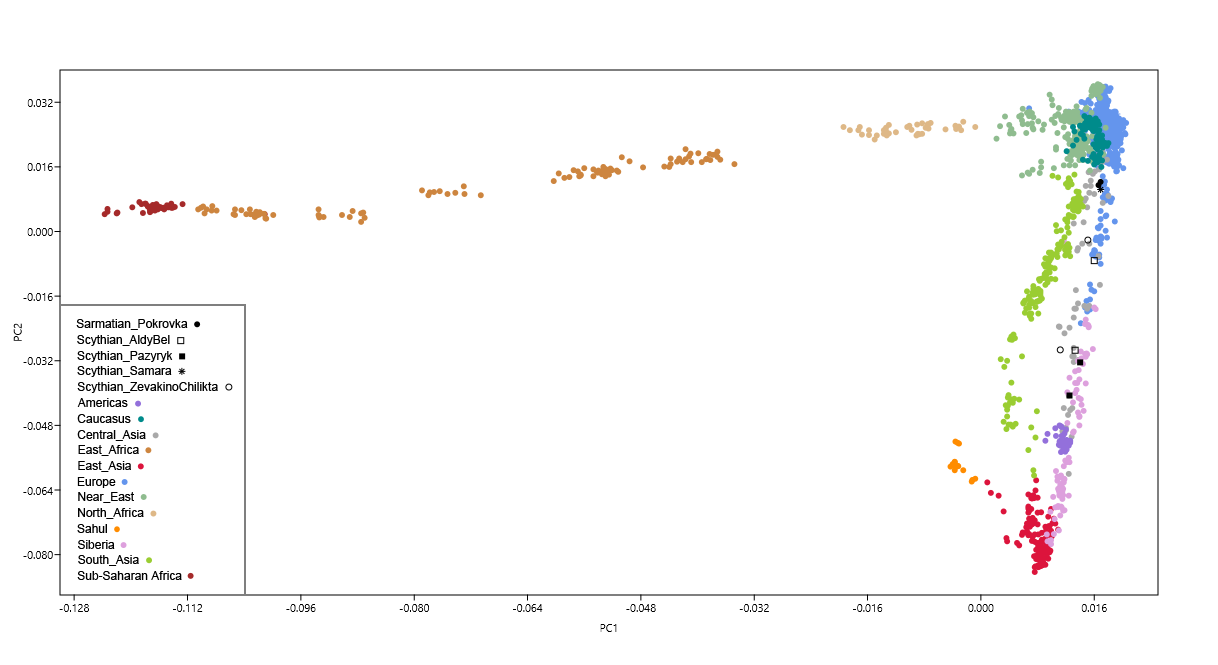 To trace genetic affinities between Scythians from present day Moldova and Ukraine (SCU) and other European and Asian ancient populations, their mt haplogroup frequencies were visualized in the space of principle components. The PCA plot of the first two components accounted for 43.4% of the total variance (Fig. 2). The SCU group was located in direct proximity to the central European Neolithic Corded Ware culture (CWC). It also grouped near a number of Bronze Age populations from eastern and central Europe (Srubnaya (SRU), Yamnaya (YAM) and Unetice (UNC)), as well as from central Asia (Bronze Age Kazakhstan (BAK)). Finally, k-means clustering (k value = 5), grouped SCU within a cluster further encompassing Scythians from Russia (SCR) and Tagar culture from southern Siberia (TAG). The Pazyryks from Mongolia (SCM) and Altai (SCA) were separated from SCU and grouped within the k-mean cluster consisting of Central and East Asian populations.  Figure 3: MDS plot based on FST calculated from complete mitochondrial genomes. Slatkin’s linearized pairwise FST values calculated on complete mt genomes and visualized using MDS (Fig. 3) and heatmap (Fig. 4) indicate that the studied Scythian samples are closest to the Srubnaya (SRU) (FST = 0.00; p < 0.05) and Yamnaya (YAM) (FST = 0.006; p < 0.05) populations (see Supplementary Table S5), followed by Unetice (UNC) (FST = 0.008; p < 0.05) and Corded Ware Culture (CWC) (FST = 0.023; p < 0.05). The European Neolithic Linear Ware culture (LBK) (FST = 0.043; p > 0.05) and Near Eastern Neolithic populations (NEN) (FST = 0.062; p > 0.05) appeared most distant to the individuals in our dataset. Correspondingly, in the resulted FST based MDS plot (stress value of 0.012), our Scythian group positioned proximately to Srubnaya (SRU), Yamnaya (YAM) and Unetice (UNC) populations and distantly to Near Eastern and European Neolithic (NEN, LBK) and Near Eastern hunter-gatherer (HGNE) populations (Fig. 3). Analysis of molecular variance (AMOVA) summarized and tested the distribution of genetic variability within and between subpopulations. We tested several other combinations of grouping Scythians with two or more populations. The results suggested the combination with the highest intragroup and lowest intergroup genetic variability to be the Scythians-Srubnaya sample-set (4.93% of variability among groups, −0.73% among populations within groups). Second best result was observed in combination of Scythians with Unetice sample-set (3.05% among groups, 1.14% among populations). The AMOVA results are summarized in Supplementary Table S6. |
|
|
|
Post by Admin on Jul 12, 2018 18:42:36 GMT
 Figure 4: Heatmap of FST and geographic distribution. Considering the high number of Scythian individuals identified as belonging to haplogroup U5 (31.6%), we conducted median network analysis of published ancient haplotypes belonging to U5a and U5b subdivisions and ranging from the Mesolithic (excluding hunter-gatherers predating last glacial maximum) to the Iron Age (Fig. 5). Haplotypes used in the median network analysis are described in detail in Supplementary Table S4. The Scythians in the U5a cluster were located centrally in the median network, near the U5a ancestral node. The U5a Scythian samples (SCU) grouped together with Mesolithic hunter-gatherers from Sweden (haplotypes 3, 4 and 5), Germany (8), Russia (10) and France (45 and 46). Non-Mesolithic samples in those clusters were less numerous and included Srubnaya (SRU) (57) and Yamnaya (YAM) (13 and 14) from Samara region and one representative of Karasuk culture from the Altai territory (central-south Siberia) (24). A single U5b Scythian individual (31) was distant from the East European populations and was placed basally to a broader cluster composed of west hunter-gatherers (HGW) from France (41) and Germany (47) as well as chronologically younger samples from the Middle Neolithic (MNE) and Unetice (UNC) (both from Germany) (15 and 53, respectively). On the basis of published data concerning the phylogeography of mt lineages distribution in ancient populations of Europe and Asia, the 19 complete mt genomes of the NPR Iron Age Scythians produced in this study fall into three main groups of different ancestry. The first group of mt lineages is represented by U5 haplotypes that are considered to be a European Hunter-Gatherer genetic component44,45. The second group comprises haplotypes belonging to H, J, T, W and N1b, ultimately connected to the genetic package of the early Neolithic farmers44,46,47, and the third group includes A, D, M10 and F mt lineages considered to be of East Eurasian origin48,49,50,51. 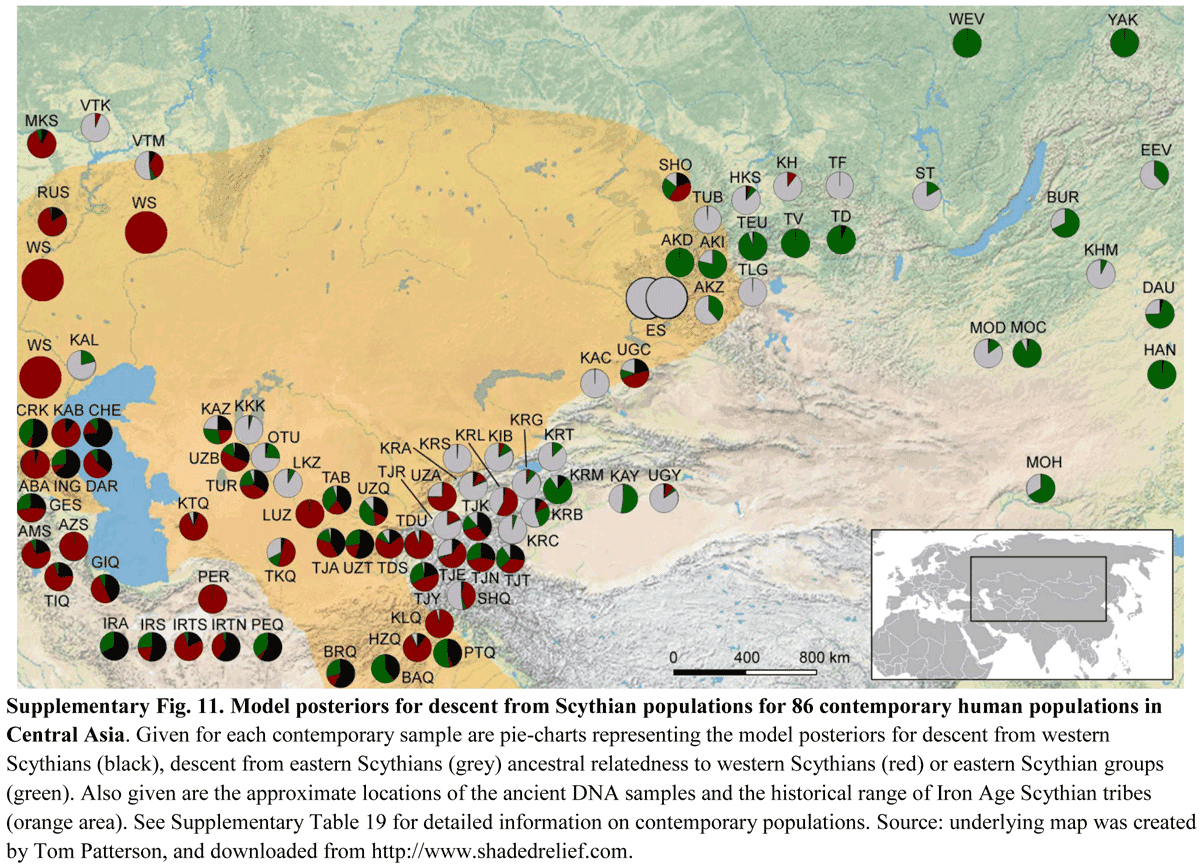 Representatives of the U5 haplogroup account for one third of the mt haplotypes identified in analyzed Scythians. These ultimately relate to West Eurasian hunter-gatherers, whose descendants extended throughout the European subcontinent and into East Eurasia from the Mesolithic to the Bronze Age. We have combined the known diversity of the prehistoric U5 lineage from different sources19,45,46,47,52,53,54,55,56 into a median network, which allowed for identification of candidate source populations contributing to the U5a diversity in NPR Scythians. Although the network approach is still limited by the small number of available samples, our results indicate that the source populations with the most closely related haplotypes are the Bronze Age Srubnaya (SRU) (1900-1200 BCE) and the earlier Yamnaya (YAM) (3300-2700 BCE) from the Ponto-Caspian region and Bronze Age populations from the Altai Mountains, such as Karasuk (1500-800 BCE). Diverse U5a-carrying populations of the steppe such as Yamnaya, Srubnaya and Scythians shared the nomadic lifestyle, with the economic foundations transforming from wild game exploitation in the Mesolithic to pastoralic animal husbandry in the Bronze Age. Furthermore, both Yamnaya and Srubnaya were part of the Kurgan culture phenomenon57 with the Scythian cultural horizon being the most representative for the kurgan building tradition. The second group of identified mt genomes in the NPR Scythians is comprised of lineages that suggest associations of the NPR Scythians with the Neolithic European farming groups. Although the mt lineage composition of analyzed Scythians significantly differs from that seen in NEN and LBK groups (FST = 0.06, FST = 0.04, respectively, p > 0.05), particular lineages such as J1c2, T2b, H/H5, H, W and N1b1a ultimately go back to the earliest European farmers. Lineages of J1c, H5 and T2b belong to the Neolithic farming package of mtDNA haplogroups which have been found in most Neolithic and Bronze Age European populations19,44,47,55,58. Noteworthy, individuals belonging to the N1b1a have been found in the Neolithic Anatolia19,59, but the lineage has not appeared in any other Eurasian Neolithic, Bronze or Iron Age populations to date. Therefore, the presence of N1b1a in Scythians could be attributed to population migrations along the southern boundaries of the Ponto-Caspian region60. |
|
|
|
Post by Admin on Nov 26, 2018 22:18:25 GMT
 Figure 5: Median network of U5 haplotypes. Representatives of the U5 haplogroup account for one third of the mt haplotypes identified in analyzed Scythians. These ultimately relate to West Eurasian hunter-gatherers, whose descendants extended throughout the European subcontinent and into East Eurasia from the Mesolithic to the Bronze Age. We have combined the known diversity of the prehistoric U5 lineage from different sources19,45,46,47,52,53,54,55,56 into a median network, which allowed for identification of candidate source populations contributing to the U5a diversity in NPR Scythians. Although the network approach is still limited by the small number of available samples, our results indicate that the source populations with the most closely related haplotypes are the Bronze Age Srubnaya (SRU) (1900-1200 BCE) and the earlier Yamnaya (YAM) (3300-2700 BCE) from the Ponto-Caspian region and Bronze Age populations from the Altai Mountains, such as Karasuk (1500-800 BCE). Diverse U5a-carrying populations of the steppe such as Yamnaya, Srubnaya and Scythians shared the nomadic lifestyle, with the economic foundations transforming from wild game exploitation in the Mesolithic to pastoralic animal husbandry in the Bronze Age. Furthermore, both Yamnaya and Srubnaya were part of the Kurgan culture phenomenon57 with the Scythian cultural horizon being the most representative for the kurgan building tradition. The second group of identified mt genomes in the NPR Scythians is comprised of lineages that suggest associations of the NPR Scythians with the Neolithic European farming groups. Although the mt lineage composition of analyzed Scythians significantly differs from that seen in NEN and LBK groups (FST = 0.06, FST = 0.04, respectively, p > 0.05), particular lineages such as J1c2, T2b, H/H5, H, W and N1b1a ultimately go back to the earliest European farmers. Lineages of J1c, H5 and T2b belong to the Neolithic farming package of mtDNA haplogroups which have been found in most Neolithic and Bronze Age European populations19,44,47,55,58. Noteworthy, individuals belonging to the N1b1a have been found in the Neolithic Anatolia19,59, but the lineage has not appeared in any other Eurasian Neolithic, Bronze or Iron Age populations to date. Therefore, the presence of N1b1a in Scythians could be attributed to population migrations along the southern boundaries of the Ponto-Caspian region60. Mitochondrial lineages T2b, W3a and J2b1a6 have been identified in representatives of the Bronze Age groups such as Srubnaya (T2b, J2b1a, H5)19, Sintashta (J2b1a, J1c)55, Yamnaya (W3a and T2b)47,61 and Mezhovskaya (J2b1a)55 along with East European Eneolithic Trypillian culture (T2b)62, suggesting genetic continuity of these lineages from at least the Bronze Age or even Neolithic times in NPR region. This is further supported by published data19 which showed that almost one fifth of the genetic makeup of the Late Bronze Age Srubnaya people of the Ponto-Caspian region is of the Early European Farmer or Anatolian Neolithic ancestry possibly resulting from the admixture of populations related to Early European Farmers and Yamnaya. Thus, if the NPR Scythians are the descendants of populations related to Srubnaya, the origin of the identified farming lineages would likely be within the steppe/forest-steppe region between the NPR and southern Ural. According to previous genomic studies47,55 the CWC people are likely to have arisen from Yamnaya background. Thus, genetic affinities of the NPR Scythians to the Yamnaya people might also explain their close genetic similarity to the CWC reflected in PCA and FST results. Close genetic relations of the NPR Scythians and Srubnaya are supported by the FST analyses revealing no significant differences between these two populations (Figs 3 and 4, Supplementary Table S5). AMOVA further corroborated these results showing that the combination of Scythians and Srubnaya results in the lowest genetic variability among populations in a group and the highest variability between groups (Supplementary Table S6). Our results support the archaeological interpretations concerning the origin of the Scythians. One of the versions of this theory suggests that the Srubnaya people have migrated in several waves from the Volga-Ural region to the NPR during the second half of the second millennium BCE where their descendants gave rise to the Scythians around the 7th century BCE63.  The Pazyryks from Mongolia and Altai were separated from the Scythians from present day Moldova and Ukraine (SCU) in the PC analysis (Figure 2) and they clustered with Central and East Asian populations (Juras et al. 2017). West Eurasian haplogroups U5 and H are dominant in the mtDNA haplogroup profile of the Scythians from Moldova and Ukraine, while the Pazyryks from Mongolia and Altai have more East Eurasian mtDNA lineages, such as A, D4j2 and F1b, compared to their SCU counterparts. East Eurasian mtDNA lineages increase from 26.3% in the Scythians (SCU) to 46.7% in the Pazyryks from Mongolia. Since haplogroup U5 accounts for 31.6% of the mtDNA haplotypes of the Scythians studied, they are genetically related to West Eurasian hunter-gatherers, whose descendants extended throughout the European subcontinent. The third group of lineages identified in the NPR Scythians is derived from East Eurasian ancestry. Since, to our knowledge, there is no evidence of agricultural subsistence in East Eurasian Scythians, these lineages should be considered to be genetic components associated with nomadic populations. Mitochondrial haplogroups such as A, D and F have already been found in samples from the Mesolithic, Neolithic, Bronze and Iron Age southern Siberia and the Altai7,14,15,16,18,55,64,65,66,67. Notably, haplogroup M10 found among Scythians from Glinoe, is present not only in far East Asia but also in modern populations of the Altai68. Among ancient populations, the M10 lineages have been found in Chinese specimens from southern Xinjiang48 (8th -1st century BCE) and Xiongnu69 (3rd century BCE). Furthermore, haplogroup M have been identified in Pleistocene individuals from western Europe but it is thought that these lineages disappeared due to bottleneck effect in the Last Glacial Maximum56. Haplogroup H8c, identified in the NPR Scythians likely belongs to East Eurasian lineages as well. Sequence analysis of 830 modern Eurasian mt genomes suggested a distinct phylogeographc history for H8, with a clustering of Near Eastern and Central Asian haplotypes of H8 and a pronounced presence of carriers of H8c in the Altai region70. It was further hypothesized that the distribution of this lineage could have been facilitated by nomadic migrations along the NPR coast70. Although our PCA analysis showed Altai Pazyryk (SCA) to be distant from the NPR Scythians (SCU) (Fig. 2), it must be emphasized that the presence of haplogroups A and H8c in the analyzed population connects NPR Scythians to the Altai and identifies this region as a possible source of these East Eurasian mt lineages. The only Scythian mt genome from southern Urals published thus far also belonged to an East Eurasian lineage, G2a19. The Y chromosomal lineage (R1a1a1b2a2a) reconstructed for this individual was supposedly characteristic for members of the Srubnaya culture19 which additionally supports our conclusions concerning close genetic links between Scythians and people related to the late Bronze Age Srubnaya. Scythians from Rostov-on-Don as well as Pazyryks from Altai and Inner Mongolia were carriers of mixed east and west Eurasian lineages, with the dominant presence of the latter at 62.5% and 53.3%, respectively7,13,14,15,16,17,18. Mitochondrial haplogroup analyses of the NPR Scythians from this study and those from Rostov-on-Don and Pazyryks from Altai and Inner Mongolia, reveal that, for the most part, the same lineages are found in all three groups and are often singularly represented in each group. Noteworthy, comparing the frequencies of east and west Eurasian haplogroups in all three groups of the Scythian horizon, an east-west mtDNA lineage cline is visible, for east Eurasian lineages going west-east is from 26.3% (in present study) through 37.5% (in Scythians from Rostov-on-Don) to 46.7% (in Pazyryks) with the opposite trend for west Eurasian lineages. Scientific Reports volume 7, Article number: 43950 (2017) |
|
|
|
Post by Admin on Aug 13, 2020 0:21:14 GMT
Y-chromosome haplogroups from Hun, Avar and conquering Hungarian period nomadic people of the Carpathian Basin Scientific Reports volume 9, Article number: 16569 (2019) Abstract Hun, Avar and conquering Hungarian nomadic groups arrived to the Carpathian Basin from the Eurasian Steppes and significantly influenced its political and ethnical landscape, however their origin remains largely unknown. In order to shed light on the genetic affinity of above groups we have determined Y chromosomal haplogroups and autosomal loci, suitable to predict biogeographic ancestry, from 49 individuals, supposed to represent the power/military elit. Haplogroups from the Hun-age are consistent with Xiongnu ancestry of European Huns. Most of the Avar-age individuals carry east Eurasian Y haplogroups typical for modern north-eastern Siberian and Buryat populations and their autosomal loci indicate mostly un-admixed Asian characteristics. In contrast the conquering Hungarians seem to be a recently assembled population incorporating un-admixed European, Asian as well as admixed components. Their heterogeneous paternal and maternal lineages indicate similar supposed phylogeographic origin of males and females, derived from Central-Inner Asian and European Pontic Steppe sources. Introduction Population history of the Carpathian Basin was profoundly determined by the invasion of various nomadic groups from the Eurasian Steppes during the Middle Ages. Between 400–453 AD the Huns held possession of the region and brought about a major population reshuffling all over Europe. Recent genetic data connect European Huns to Inner Asian Xiongnus1, but genetic data from Huns of the Carpathian Basin have not been available yet, since Huns left just sporadic lonely graves in the region, as they stayed for short period. We report three Y haplogroups (Hg) from Hun age remains, which possibly belonged to Huns based on their archaeological and anthropological evaluation.  From 568 AD the Avars established an empire in the region lasting nearly for 250 years, until they were defeated by the Franks and Bulgars in 803, then their steppe-empire ended around 822 AD. In its early stage the Avar Khaganate controlled a large territory expanding from the Carpathian Basin to the Pontic-Caspian Steppes and dominated numerous folks including Onogur-Bulgars, which fought their independence in the middle 7th century and established the independent Magna Bulgaria state. Then the Avar Khaganate was shrunken, its range well corresponding to that of the succeeding Hungarian Kingdom. The Avars arrived in multiple waves into the Carpathian Basin and the Avar period left a vast archeological legacy with more than 80 thousand excavated graves in present-day Hungary. The Avar age remains are anthropologically extremely heterogeneous, with considerable proportion of Mongoloid and Europo-Monoloid elements reaching 20–30% on the Great Hungarian Plain2, attesting that the Carpathian Basin witnessed the largest invasion of people from Asia during this period. Most individuals buried with rich grave goods show Mongoloid characters indicating inner Asian origin of the Avar elite, which is also supported by their artifact types, titles (e.g. khagan) and institutions recognized to be derived from Inner Asian Rourans. From the Avar period only a few mitochondrial DNA (mtDNA) data are available from two micro-regions3,4, which showed 15.3% and 6.52% frequency of East Eurasian elements. A recent manuscript described 23 mitogenomes from the 7th-8th century Avar elite group5 and found that 64% of the lineages belong to East Asian haplogroups (C, D, F, M, R, Y and Z) with affinities to ancient and modern Inner Asian populations corroborating their Rouran origin. Though the Avar Khaganate ceased to exist around 822 AD, but its population survived and were incorporated into the succeeding Hungarian state6. It is relevant to note that none of the Hungarian medieval sources know about Avars7, probably because they were not distinguished from the Huns as many foreign medieval sources also identified Avars with Huns, for example the Avars who were Christianized and became tax-payer vassals of the Eastern Frankish Empire were called as Huns in 8718. Presence of the Hungarians in the Carpathian Basin was documented from 862 AD and between 895–905 they took full command of the region. The Hungarians formed a tribal union but arrived in the frame of a strong centralized steppe-empire under the leadership of prince Álmos and his son Árpád, who were known to be direct descendants of the great Hun leader Attila, and became founders of the Hungarian ruling dynasty and the Hungarian state. The Hungarian Great Principality existed in Central Europe from ca. 862 until 1000, then it was re-organized as a Christian Kingdom by King István I the Saint who was the 5th descendant of Álmos9. Our recent analysis of conquering Hungarian (hence shortened as Conqueror) mitogenomes revealed that the origin of their maternal lineages can be traced back to distant parts of the Eurasian steppe10. One third of the maternal lineages were derived from Central-Inner Asia and their most probable ultimate sources were the Asian Scythians and Asian Huns, while the majority of the lineages most likely originated from the Bronze Age Potapovka-Poltavka-Srubnaya cultures of the Pontic-Caspian steppe. Population genetic analysis indicated that Conquerors had closest connection to the Onogur-Bulgar ancestors of Volga Tatars. Although mtDNA data can be informative, since as far as we know none of the above nomadic invasions were pure military expeditions in which raiding males took local women, but entire societies with both men and women migrated together3, however nomadic societies were patrilinealy organized, thus Y-chromosome data are expected to provide more relevant information about their structure and origin than mtDNA. So far 6 Y-chromosome Hg-s have been published from the Conquerors11; which revealed the presence of N1a1- M46 (previously called Tat or N1c), in two out of 4 men, while12 detected two R1b-U106 and two I2a-M170 Hg-s. In order to generate sufficient data for statistical evaluation and compare paternal and maternal lineages of the same population, we have determined Y-Hg-s from the same cemeteries whose mtDNA Hg-s were described in10. As most Hungarian medieval chronicles describe the conquer as the “second incoming of the Hungarians”7, genetic relations may exist between consecutive nomadic groups, first of all between elite groups. As the studied Conqueror cemeteries represent mainly the Conqueror elite, we supplemented our Y-Hg studies with samples from the preceding Avar power elite/millitary elite groups and available samples from the Hun period to test possible genetic relations. Avar military leader graves are distinguished by high-value prestige artifacts: belt decorated with gold or silver mounts, decorated horse harness, saddle, sword, bow and quiver with arrowheads, gold decorated plates, silver or gold earrings (Supplementary Information). Conqueror leader graves contain characteristic partial horse burial with horse cranium and leg bones or symbolic horse burial with adorned harness, saddle and stirrup. Elite grave goods often include precious metal jewels, belt with impressed metal belt-buckles, metal-plated sabretache and weaponry; arrowheads, quiver and bow10,12. From the studied samples we also tested autosomal single nucleotide polymorphisms (SNP-s) associated with eye/hair/skin color and lactase persistence phenotypes as well as biogeographic ancestry. |
|
|
|
Post by Admin on Aug 13, 2020 8:11:48 GMT
Results We selected 168 phylogenetically informative Y chromosome SNP-s13 defining all major Hg-s and the most frequent Eurasian sub-Hg-s, as well as the following 61autosomal SNP-s: 25 HirisPlex markers suitable for eye and hair color prediction14, two SNP-s linked to adult lactase persistence15 and 34 ancestry informative markers (AIMs)16, as listed in Supplementary Tables S1 and S2. DNA segments containing above SNP-s were enriched from the next generation sequencing (NGS) library of each sample with hybridization capture. Anthropological males were selected from the cemeteries, but as control we also included four females for which the presence of Y-chromosomal reads can reveal contamination. Analysis of women control samples revealed low contamination, having negligible Y-chromosomal reads but comparable autosomal reads to males (Supplementary Tables S1 and S2) with one exception, KEF1/10936 genetically turned out to be a male despite its anthropological description as female (Supplementary Table S6). Negligible contamination levels for 26 of the 49 libraries had been demonstrated before10 and low contamination is also inferred from unambiguous Hg classifications with very few contradicting SNP-s, most of which can be explained by postmortem ancient DNA modifications (Supplementary Table S1). Besides, transition patterns of DNA molecules and library fragment size distributions were typical for ancient DNA and we also estimated low contamination by calculating the proportion of reads which did not correspond to the consensus sequence in diagnostic positions (Supplementary Table S3). Moreover we detected a significant number of east Eurasian Hg-s which had a negligible chance to have been derived from a contaminating source of European researchers. We obtained low to medium average coverage of the targeted loci, (Supplementary Tables S1, S2 and S3) and Y-Hg determination was consistent from 46 males (Supplementary Table S1) which is summarized in Fig. 1 and phylogeographic distribution of Conqueror lineages is displayed on Fig. 2. Figure 1 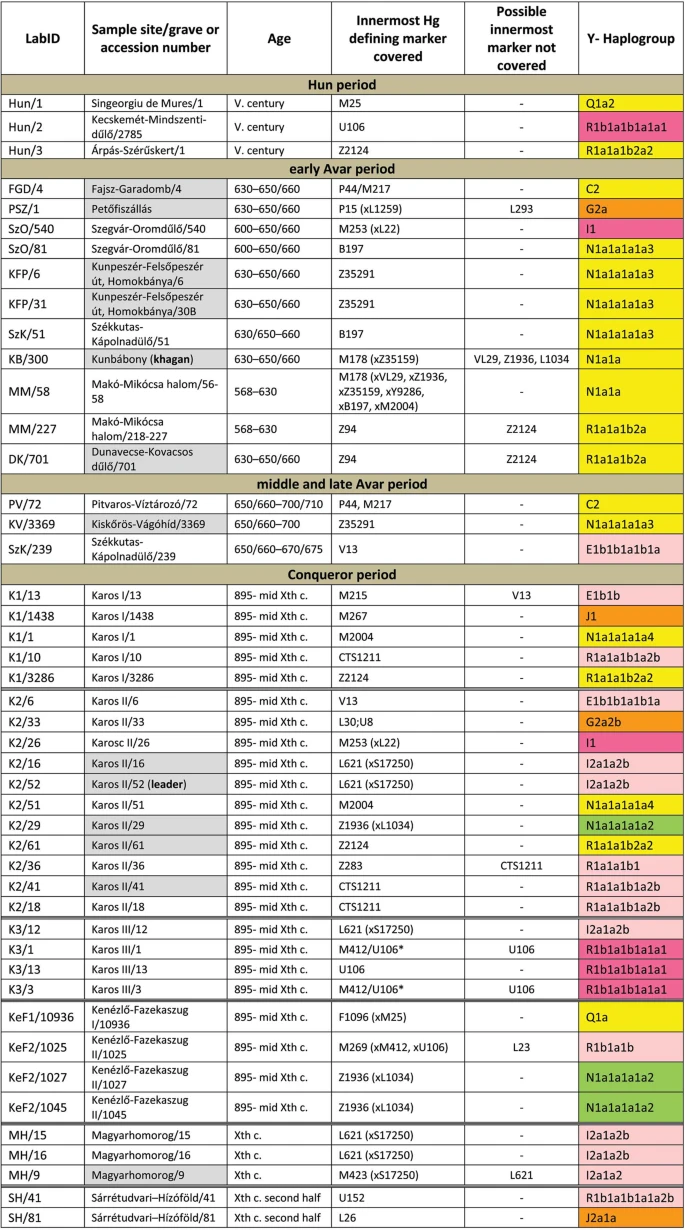 East Eurasian Hg-s Hg Q1a2- M25 is very rare in Europe, where it has highest frequency among Seklers (a Hungarian speaking ethnic group in Transylvania) according to Family Tree DNA database. Ancient samples with Hg Q1a2- M25 are known from the Bronze Age Okunevo and Karasuk cultures, as well as Middle Age Tian Shan Huns and Hunnic-Sarmatians17 implying possible Hunnic origin of this lineage in Europe, which is confirmed by the Hg of our Hun/1 sample, derived from Transylvania. One Conqueror sample KEF1/10936 belongs to Q1a- F1096, possibly to the Q1a1-F746 sister clade of Q1a2- M25, which was not tested in our experiment. Q1a1 is present in East Asia; Mongolia, Japan, China, Korea and its presence in the Kenézlő (KEF) graveyard imply a common substrate of Huns and Conquerors. Hun/3 belongs to Hg R1a1a1b2a2- Z2124, a subclade of R1a1a1b2-Z93, the east Eurasian subbranch of R1a. Today Z2124 is most frequent in Kyrgyzstan and Afghanistan, but is also widespread among Karachai-Balkars and Baskhirs18. Z2124 was widespread on the Bronze Age steppe, especially in the Afanasievo and Sintashta cultures19 and R1a detected in Xiongnus20,21 very likely belong to the same branch. Two samples from the Karos Conqueror cemeteries (K1/3286 and K2/61) were also classified as R1a-Z2124 and two Avar age individuals (DK/701 and MM/227) belong to the same R1a1a1b2a-Z94 branch but marker Z2124 was not covered in latter samples. Two Avar samples belonged to Hg C2-M217, which is found mostly in Central Asia and Eastern Siberia. Presence of this Hg had also been detected in the Conquerors, as the Karos2/60 individual belongs to C2 (Fóthi unpublished). All N-Hg-s identified in the Avars and Conquerors belonged to N1a1a-M178. We have tested 7 subclades of M178; N1a1a2-B187, N1a1a1a2-B211, N1a1a1a1a3-B197, N1a1a1a1a4-M2118, N1a1a1a1a1a-VL29, N1a1a1a1a2-Z1936 and the N1a1a1a1a2a1c1-L1034 subbranch of Z1936. The European subclades VL29 and Z1936 could be excluded in most cases, while the rest of the suclades are prevalent in Siberia22 from where this Hg dispersed in a counter-clockwise migratory route to Europe23. Avar sample MM/58, did not go into any of the tested M178 subclades, while only N1a1a2 could be excluded for the KB/300 Avar khagan due to low coverage. All the 5 other Avar samples belonged to N1a1a1a1a3-B197, which is most prevalent in Chukchi, Buryats, Eskimos, Koryaks and appears among Tuvans and Mongols with lower frequency22. By contrast two Conquerors belonged to N1a1a1a1a4-M2118, the Y lineage of nearly all Yakut males, being also frequent in Evenks, Evens and occurring with lower frequency among Khantys, Mansis and Kazakhs. Figure 2 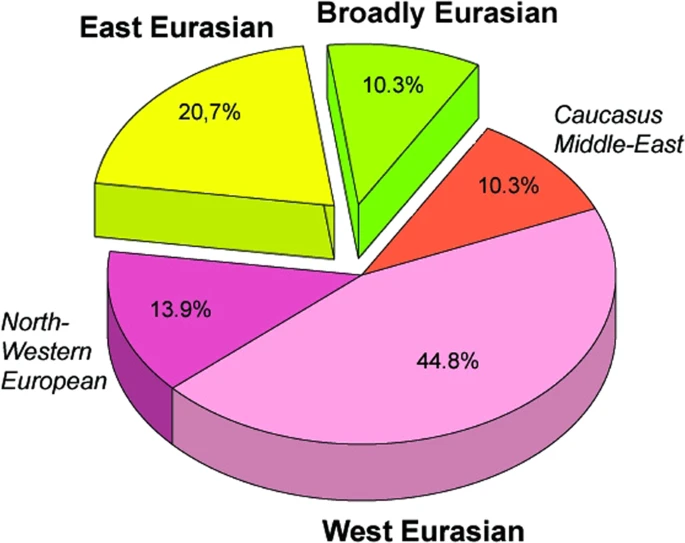 Broadly Eurasian Hg-s Three Conqueror samples belonged to Hg N1a1a1a1a2-Z1936, the Finno-Permic N1a branch, being most frequent among northeastern European Saami, Finns, Karelians, as well as Komis, Volga Tatars and Bashkirs of the Volga-Ural region. Nevertheless this Hg is also present with lower frequency among Karanogays, Siberian Nenets, Khantys, Mansis, Dolgans, Nganasans, and Siberian Tatars22. West Eurasian Hg-s The west Eurasian R1a1a1b1a2b-CTS1211 subclade of R1a is most frequent in Eastern Europe especially among Slavic people. This Hg was detected just in the Conqueror group (K2/18, K2/41 and K1/10). Though CTS1211 was not covered in K2/36 but it may also belong to this sub-branch of Z283. Hg I2a1a2b-L621 was present in 5 Conqueror samples, and a 6th sample form Magyarhomorog (MH/9) most likely also belongs here, as MH/9 is a likely kin of MH/16 (see below). This Hg of European origin is most prominent in the Balkans and Eastern Europe, especially among Slavic speaking groups. It might have been a major lineage of the Cucuteni-Trypillian culture and it was present in the Baden culture of the Calcholitic Carpathian Basin24. I1- M253, identified from one Conqueror sample is a northern European Hg found mostly in Scandinavia and Finland and might have originated from this region during the Mesolithic. It has somewhat similar distribution to R1b-U106 associated with Germanic speaking populations. Three out of 4 samples in the small Karos3 cemetery belonged to Hg R1b1a1b1a1a1-U106 setting apart this cemetery from all other groups, except for the Hun/2 sample which is the only other one with this Hg. Hg U106 is considered a “Germanic” branch as it is most significant today in Germany, Scandinavia, and Britain, and rare in Eastern Europe (Supplementary Table S4). Its ancestral branch Hg R1b1a1b-M262 is assumed to have emerged in the Pontic-Caspian Steppe and arrived to Europe with Bronze Age migrations25. Its presence in Hun and Conqueror samples may derive from Goths, Gepids or other German allies of the Huns. We detected R1b1a1b1a1a2b-U152 in one sample from the Sárrétudvari Conqueror cemetery, which represent rather commoners than the elite. U152 is the Italo-Celtic R1b branch, concentrated around the Alps, and which was present in the Carpathian Basin before the conquer, so did not necessarily arrived with the Conquerors. The mediterranean haplogroup E1b1b1a1b1a-V13 was detected in an Avar (SzK/239) and a Conqueror (K2/6) sample, while this marker was not covered in another sample (K1/13, E1b1b- M215). This Hg originated in the Middle East and migrated to the Balkans and Western Asia during the Bronze Age. The PSZ/1 Avar leader belongs to Hg G2a-P15, while K2/33 to the G2a2b-L30 subbranch. Hg G2a is originated from Anatolia/Iran, now it is most common in the Caucasus region and its arrival to Europe is associated with the spread of Neolithic farmers26. One Conqueror sample belongs to Hg J1-M267 and another to J2a1a-L26. Both J1 and J2 lineages are most frequent around the Middle East-Caucasus and probably originated from this region27, then expanded with pastorists prior to the Neolithic. |
|










